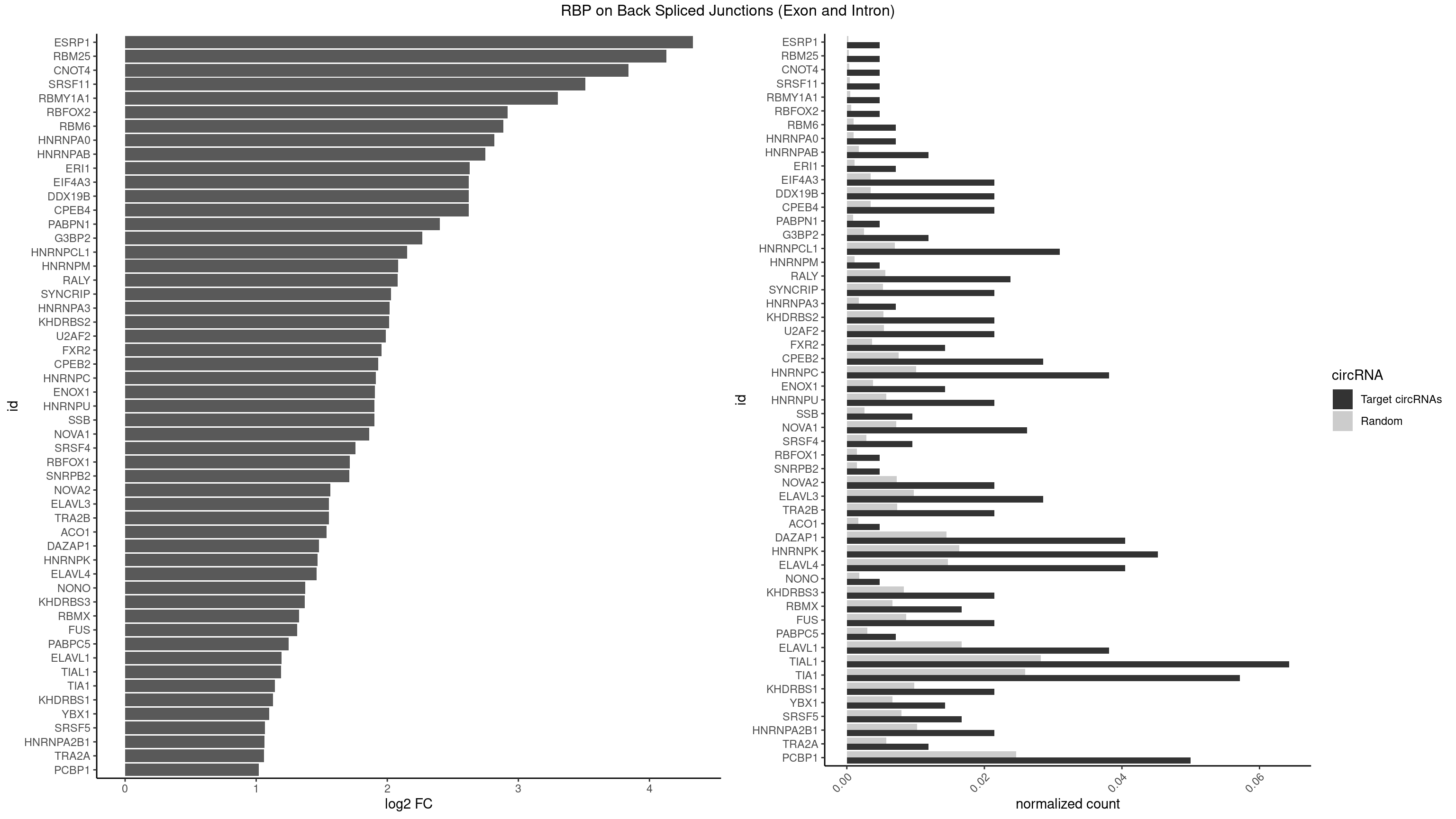
| ESRP1 |
1 |
46 |
0.004761905 |
0.0002367997 |
4.329800 |
AGGGAU |
AGGGAU |
| RBM25 |
1 |
53 |
0.004761905 |
0.0002720677 |
4.129501 |
CGGGCA |
AUCGGG,CGGGCA,UCGGGC |
| CNOT4 |
1 |
65 |
0.004761905 |
0.0003325272 |
3.839994 |
GACAGA |
GACAGA |
| SRSF11 |
1 |
82 |
0.004761905 |
0.0004181782 |
3.509349 |
AAGAAG |
AAGAAG |
| RBMY1A1 |
1 |
95 |
0.004761905 |
0.0004836759 |
3.299426 |
ACAAGA |
ACAAGA,CAAGAC |
| RBFOX2 |
1 |
124 |
0.004761905 |
0.0006297864 |
2.918604 |
UGACUG |
UGACUG,UGCAUG |
| RBM6 |
2 |
191 |
0.007142857 |
0.0009673519 |
2.884389 |
AAUCCA,AUCCAG |
AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPA0 |
2 |
200 |
0.007142857 |
0.0010126965 |
2.818299 |
AAUUUA,AGAUAU |
AAUUUA,AGAUAU,AGUAGG |
| HNRNPAB |
4 |
351 |
0.011904762 |
0.0017734784 |
2.746885 |
AAGACA,ACAAAG,AGACAA,CAAAGA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ERI1 |
2 |
228 |
0.007142857 |
0.0011537686 |
2.630147 |
UUCAGA,UUUCAG |
UUCAGA,UUUCAG |
| CPEB4 |
8 |
691 |
0.021428571 |
0.0034864974 |
2.619685 |
UUUUUU |
UUUUUU |
| DDX19B |
8 |
691 |
0.021428571 |
0.0034864974 |
2.619685 |
UUUUUU |
UUUUUU |
| EIF4A3 |
8 |
691 |
0.021428571 |
0.0034864974 |
2.619685 |
UUUUUU |
UUUUUU |
| PABPN1 |
1 |
178 |
0.004761905 |
0.0009018541 |
2.400573 |
AAAAGA |
AAAAGA,AGAAGA |
| G3BP2 |
4 |
490 |
0.011904762 |
0.0024738009 |
2.266737 |
AGGAUG,GGAUGA,GGAUUA,GGAUUG |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPCL1 |
12 |
1381 |
0.030952381 |
0.0069629182 |
2.152286 |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| HNRNPM |
1 |
222 |
0.004761905 |
0.0011235389 |
2.083489 |
AAGGAA |
AAGGAA,GAAGGA,GGGGGG |
| RALY |
9 |
1117 |
0.023809524 |
0.0056328094 |
2.079612 |
UUUUUG,UUUUUU |
UUUUUC,UUUUUG,UUUUUU |
| SYNCRIP |
8 |
1041 |
0.021428571 |
0.0052498992 |
2.029174 |
UUUUUU |
AAAAAA,UUUUUU |
| HNRNPA3 |
2 |
349 |
0.007142857 |
0.0017634019 |
2.018140 |
AGGAGC,GGAGCC |
AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| KHDRBS2 |
8 |
1051 |
0.021428571 |
0.0053002821 |
2.015395 |
UUUUUU |
AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| U2AF2 |
8 |
1071 |
0.021428571 |
0.0054010480 |
1.988224 |
UUUUUU |
UUUUCC,UUUUUC,UUUUUU |
| FXR2 |
5 |
730 |
0.014285714 |
0.0036829907 |
1.955624 |
AGACAA,AGACAG,GACAAG,GACAGA,GGACAG |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| CPEB2 |
11 |
1487 |
0.028571429 |
0.0074969770 |
1.930192 |
AUUUUU,CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPC |
15 |
2006 |
0.038095238 |
0.0101118501 |
1.913564 |
AUUUUU,CUUUUU,UUUUUA,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ENOX1 |
5 |
756 |
0.014285714 |
0.0038139863 |
1.905202 |
AAGACA,AGACAG,AGGACA,CAGACA,GGACAG |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPU |
8 |
1136 |
0.021428571 |
0.0057285369 |
1.903297 |
UUUUUU |
AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| SSB |
3 |
505 |
0.009523810 |
0.0025493753 |
1.901395 |
CUGUUU,GCUGUU,UGUUUU |
CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| NOVA1 |
10 |
1428 |
0.026190476 |
0.0071997179 |
1.863030 |
CAGUCA,UCAGUC,UUUUUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| SRSF4 |
3 |
558 |
0.009523810 |
0.0028164047 |
1.757684 |
AAGAAG,AGGAAG,GAGGAA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| RBFOX1 |
1 |
287 |
0.004761905 |
0.0014510278 |
1.714464 |
UGACUG |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 |
1 |
288 |
0.004761905 |
0.0014560661 |
1.709463 |
GUAUUG |
AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| NOVA2 |
8 |
1434 |
0.021428571 |
0.0072299476 |
1.567479 |
UUUUUU |
AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| ELAVL3 |
11 |
1928 |
0.028571429 |
0.0097188634 |
1.555714 |
AUUUAU,UAUUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| TRA2B |
8 |
1447 |
0.021428571 |
0.0072954454 |
1.554468 |
AAAGAA,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGG,AGGAAA,AGGAAG |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| ACO1 |
1 |
325 |
0.004761905 |
0.0016424829 |
1.535660 |
CAGUGU |
CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| DAZAP1 |
16 |
2878 |
0.040476190 |
0.0145052398 |
1.480499 |
AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUG,AGUAAA,AGUAAG,UAGGAU,UUUUUU |
AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPK |
18 |
3244 |
0.045238095 |
0.0163492543 |
1.468313 |
ACCCCA,ACCCCC,CCCCAC,CCCCAU,CCCCCA,GCCCAA,GCCCAG,UCCCAC,UCCCAU,UCCCCA,UUUUUU |
AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| ELAVL4 |
16 |
2916 |
0.040476190 |
0.0146966949 |
1.461582 |
AUUUAU,UAUUUU,UUUAUU,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| NONO |
1 |
364 |
0.004761905 |
0.0018389762 |
1.372636 |
GAGGAA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| KHDRBS3 |
8 |
1646 |
0.021428571 |
0.0082980653 |
1.368689 |
UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| RBMX |
6 |
1316 |
0.016666667 |
0.0066354293 |
1.328704 |
AAGAAG,AAGGAA,AAGUAA,AGAAGG,AGGAAG,AUCCCA |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| FUS |
8 |
1711 |
0.021428571 |
0.0086255542 |
1.312847 |
UUUUUU |
AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| PABPC5 |
2 |
596 |
0.007142857 |
0.0030078597 |
1.247764 |
AGAAAU,GAAAUU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ELAVL1 |
15 |
3309 |
0.038095238 |
0.0166767432 |
1.191773 |
AUUUAU,UAUUUU,UGUUUU,UUGUUU,UUUAUU,UUUGUU,UUUUUG,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| TIAL1 |
26 |
5597 |
0.064285714 |
0.0282043531 |
1.188580 |
AAAUUU,AUUUUU,CAGUUU,CUUUUU,GUUUUC,GUUUUU,UAUUUU,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUG,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TIA1 |
23 |
5140 |
0.057142857 |
0.0259018541 |
1.141518 |
AUUUUU,CUUUUU,GUUUUC,GUUUUU,UAUUUU,UCUCCU,UUUAUU,UUUUAU,UUUUGU,UUUUUA,UUUUUG,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS1 |
8 |
1945 |
0.021428571 |
0.0098045143 |
1.128018 |
UUUUUU |
AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| YBX1 |
5 |
1321 |
0.014285714 |
0.0066606207 |
1.100845 |
ACCACA,CAGCAA,CCAGCA,GAUCUG,UCCAGC |
AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| SRSF5 |
6 |
1578 |
0.016666667 |
0.0079554615 |
1.066948 |
AAGAAG,AGGAAG,CACAGC,GAGGAA,UGCAGA,UGCAGC |
AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| HNRNPA2B1 |
8 |
2033 |
0.021428571 |
0.0102478839 |
1.064210 |
AAGAAG,AAGGAA,AAUUUA,AGAUAU,AGGAGC,CAAGAA,CAAGGG,GGAGCC |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| TRA2A |
4 |
1133 |
0.011904762 |
0.0057134220 |
1.059112 |
AAAGAA,AAGAAG,AGGAAG,GAGGAA |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PCBP1 |
20 |
4891 |
0.050000000 |
0.0246473196 |
1.020497 |
ACCCUC,AUUACC,CACCCU,CCAUUA,CCAUUC,CCCCAC,CCCUCU,CCUAAC,CCUCUU,CUUUCC,UCCCCA,UUUUUU |
AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |