circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000048828:+:9:93471140:93476338
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000048828:+:9:93471140:93476338 | ENSG00000048828 | ENST00000277165 | + | 9 | 93471141 | 93476338 | 330 | GUUGCACAGAGCAUUGAGGAUCACCAUCAGGAAGUGAUUGGUUUCUGCAGAGAGAAUGGUUUCCAUGGCUUGGUUGCGUAUGACUCUGAUUAUGCACUGUGCAACAUCCCCUACUAUUUCAGUGCCCAUGCCCUAAAACUGAGCCGGAACGGGAAAAGUCUCACCACAAGCCAAUAUCUGAUGCAUGAAGUUGCCAAGCAACUGGACCUGAAUCCAAAUCGUUUUCCUAUUUUUGCUGCUCUCUUAGGAAAUCACAUUCUGCCUGAUGAAGAUCUGGCUUCCUUUCACUGGAGUUUACUUGGUCCAGAACAUCCACUAGCCUCACUAAAGGUUGCACAGAGCAUUGAGGAUCACCAUCAGGAAGUGAUUGGUUUCUGCAG | circ |
| ENSG00000048828:+:9:93471140:93476338 | ENSG00000048828 | ENST00000277165 | + | 9 | 93471141 | 93476338 | 22 | CCUCACUAAAGGUUGCACAGAG | bsj |
| ENSG00000048828:+:9:93471140:93476338 | ENSG00000048828 | ENST00000277165 | + | 9 | 93470941 | 93471150 | 210 | UUUAAAUGCGAUUUCCCACAGAAAUUUCUUAGUGGAGCAAUGUGUUGCAAACUUUAUUAUGGCAUUUUUUUGACUGUUUGGCUUUCACGGAUAGUUUUCAUAAAUGUGAUUUUUCAGCUUCUGUGCAAACAUUUCUGCUUAUUGAGCUUGUUGCUACAGUGUUACCCUACAUCUGAUGAAGUUUUUUUCUCGUUUGCCAGGUUGCACAGA | ie_up |
| ENSG00000048828:+:9:93471140:93476338 | ENSG00000048828 | ENST00000277165 | + | 9 | 93476329 | 93476538 | 210 | CUCACUAAAGGUACAAAUUUCACUUUAUUUUUCUAGCAUUUGUAAUAAUCAACUGUGAGUUUGCUAUUACUUUAAUAACAUGUUAGGCCCUUUAUUGGUGAUGCUACAAAGUUAGCAAUACCAUGUUUUCCCAUAAUUUCAGGAUCUGCACUGCCUUAAAAAGCAAGCAACAAUGCUACCAUCCCUUGUUCUCAACACCUAUAUGUUAGG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
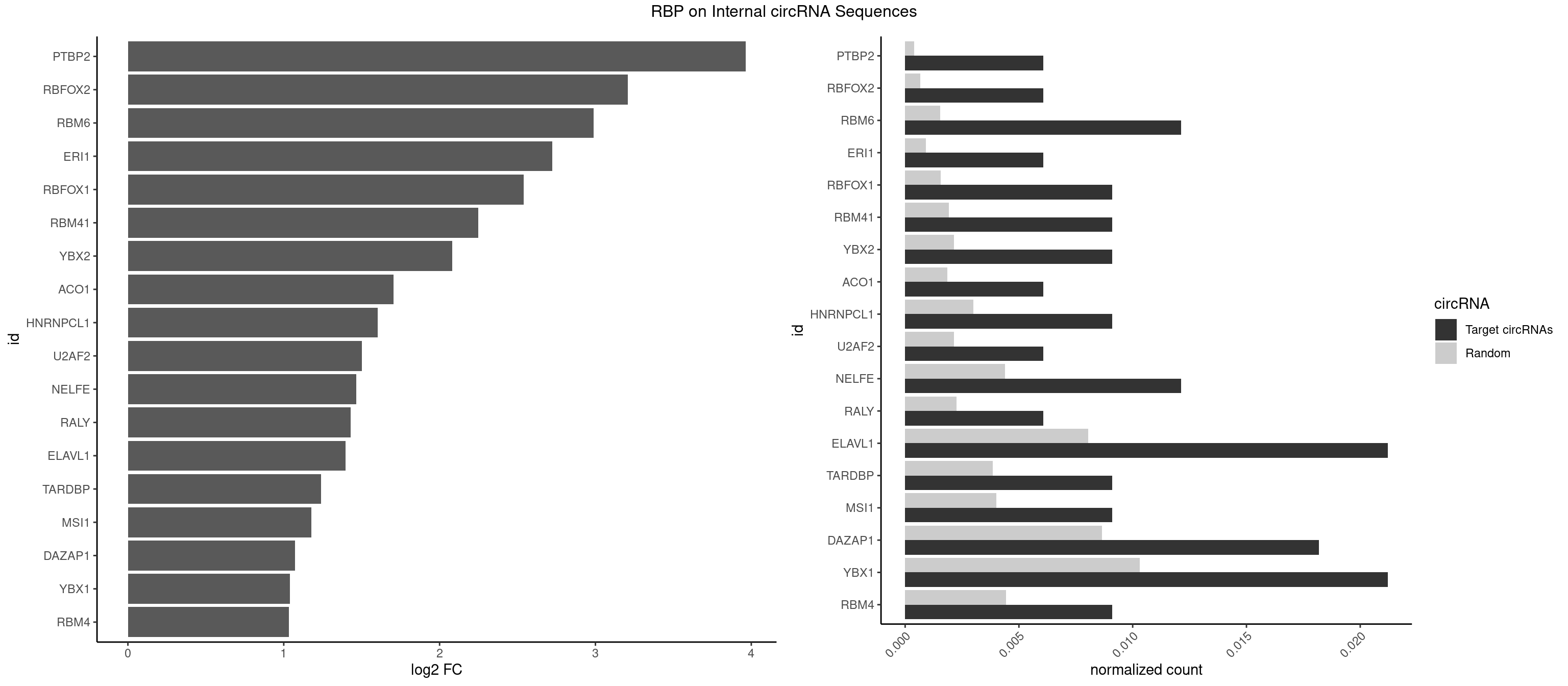
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 267 | 0.006060606 | 0.0003883867 | 3.963897 | CUCUCU | CUCUCU |
| RBFOX2 | 1 | 452 | 0.006060606 | 0.0006564894 | 3.206618 | UGCAUG | UGACUG,UGCAUG |
| RBM6 | 3 | 1054 | 0.012121212 | 0.0015289102 | 2.986958 | AAUCCA,AUCCAA,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ERI1 | 1 | 632 | 0.006060606 | 0.0009173461 | 2.723924 | UUUCAG | UUCAGA,UUUCAG |
| RBFOX1 | 2 | 1077 | 0.009090909 | 0.0015622419 | 2.540807 | GCAUGA,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBM41 | 2 | 1318 | 0.009090909 | 0.0019115000 | 2.249719 | UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| YBX2 | 2 | 1480 | 0.009090909 | 0.0021462711 | 2.082592 | AACAUC | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| ACO1 | 1 | 1283 | 0.006060606 | 0.0018607779 | 1.703556 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPCL1 | 2 | 2062 | 0.009090909 | 0.0029897078 | 1.604420 | AUUUUU,UUUUUG | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| U2AF2 | 1 | 1477 | 0.006060606 | 0.0021419234 | 1.500555 | UUUUCC | UUUUCC,UUUUUC,UUUUUU |
| NELFE | 3 | 3028 | 0.012121212 | 0.0043896388 | 1.465360 | CUCUCU,CUGGCU,UCUGGC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RALY | 1 | 1553 | 0.006060606 | 0.0022520629 | 1.428215 | UUUUUG | UUUUUC,UUUUUG,UUUUUU |
| ELAVL1 | 6 | 5554 | 0.021212121 | 0.0080503280 | 1.397769 | UAUUUU,UGGUUU,UUGGUU,UUUUUG | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| TARDBP | 2 | 2654 | 0.009090909 | 0.0038476365 | 1.240452 | GAAUGG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| MSI1 | 2 | 2770 | 0.009090909 | 0.0040157442 | 1.178757 | AGGAAG,UAGGAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| DAZAP1 | 5 | 5964 | 0.018181818 | 0.0086445016 | 1.072642 | AGGAAA,AGGAAG,AGGUUG,AGUUUA,UAGGAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| YBX1 | 6 | 7119 | 0.021212121 | 0.0103183321 | 1.039679 | AACAUC,ACCACA,CACCAC,CCACAA,GAUCUG | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| RBM4 | 2 | 3065 | 0.009090909 | 0.0044432593 | 1.032806 | CUUCCU,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
RBP on BSJ (Exon Only)
Plot
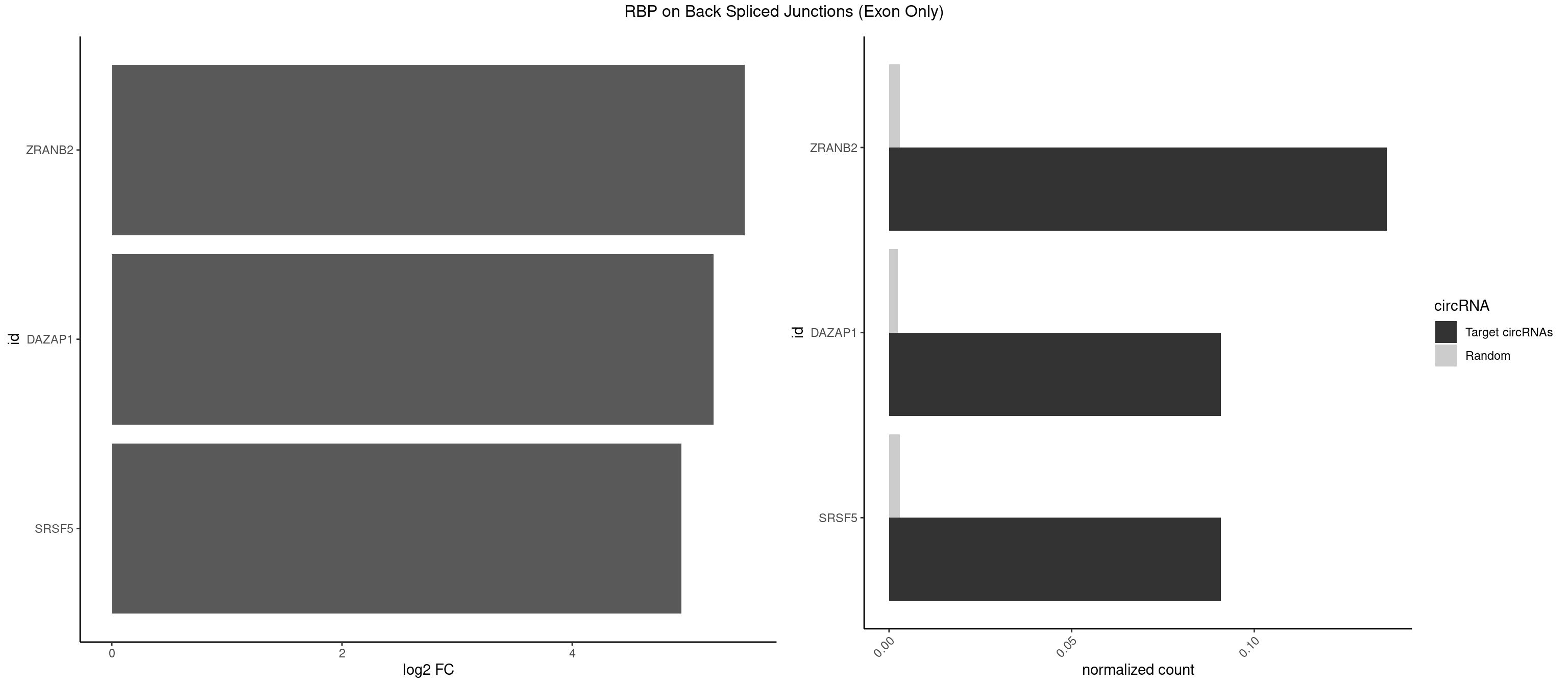
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZRANB2 | 2 | 45 | 0.13636364 | 0.003012837 | 5.500192 | AAAGGU,UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.002423369 | 5.229338 | AGGUUG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| SRSF5 | 1 | 44 | 0.09090909 | 0.002947341 | 4.946939 | UAAAGG | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUC,CACGGA,CGCAGC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACGUA,UGCAGA,UGCAGC,UGCGGC |
RBP on BSJ (Exon and Intron)
Plot
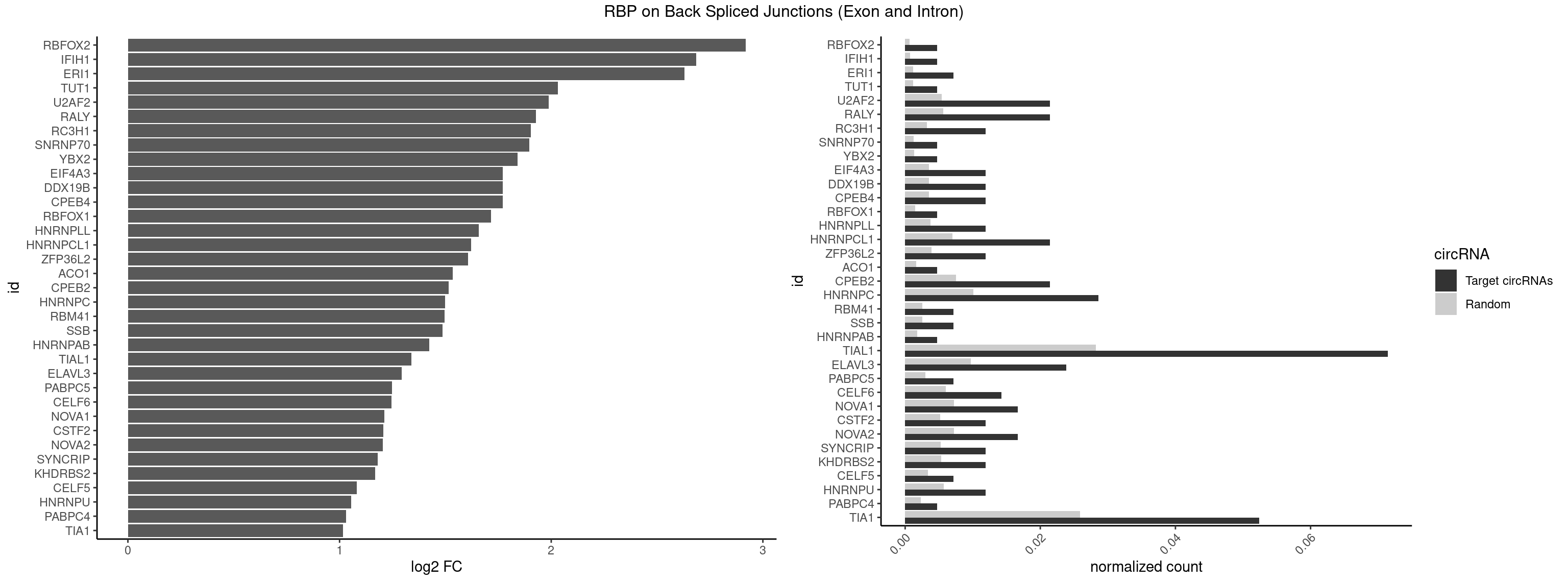
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| U2AF2 | 8 | 1071 | 0.021428571 | 0.0054010480 | 1.988224 | UUUUCC,UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RALY | 8 | 1117 | 0.021428571 | 0.0056328094 | 1.927609 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| RC3H1 | 4 | 631 | 0.011904762 | 0.0031841999 | 1.902536 | CUUCUG,UCCCUU,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CPEB4 | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| DDX19B | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| EIF4A3 | 4 | 691 | 0.011904762 | 0.0034864974 | 1.771688 | UUUUUU | UUUUUU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | CAAACA,CACUGC,GCAAAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| HNRNPCL1 | 8 | 1381 | 0.021428571 | 0.0069629182 | 1.621772 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| ZFP36L2 | 4 | 774 | 0.011904762 | 0.0039046755 | 1.608264 | UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CPEB2 | 8 | 1487 | 0.021428571 | 0.0074969770 | 1.515155 | AUUUUU,CAUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPC | 11 | 2006 | 0.028571429 | 0.0101118501 | 1.498526 | AUUUUU,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| TIAL1 | 29 | 5597 | 0.071428571 | 0.0282043531 | 1.340583 | AAAUUU,AGUUUU,AUUUUU,GUUUUC,GUUUUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUAUU,UUUCAG,UUUUCA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL3 | 9 | 1928 | 0.023809524 | 0.0097188634 | 1.292679 | UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| CELF6 | 5 | 1196 | 0.014285714 | 0.0060308343 | 1.244144 | GUGAUG,GUGUUG,UGUGAG,UGUGAU,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| NOVA1 | 6 | 1428 | 0.016666667 | 0.0071997179 | 1.210953 | AACACC,AUCAAC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUUG,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| NOVA2 | 6 | 1434 | 0.016666667 | 0.0072299476 | 1.204908 | AACACC,AUCAAC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| SYNCRIP | 4 | 1041 | 0.011904762 | 0.0052498992 | 1.181177 | UUUUUU | AAAAAA,UUUUUU |
| KHDRBS2 | 4 | 1051 | 0.011904762 | 0.0053002821 | 1.167398 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPU | 4 | 1136 | 0.011904762 | 0.0057285369 | 1.055300 | UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
| TIA1 | 21 | 5140 | 0.052380952 | 0.0259018541 | 1.015987 | AUUUUU,GUUUUC,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUCU,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.