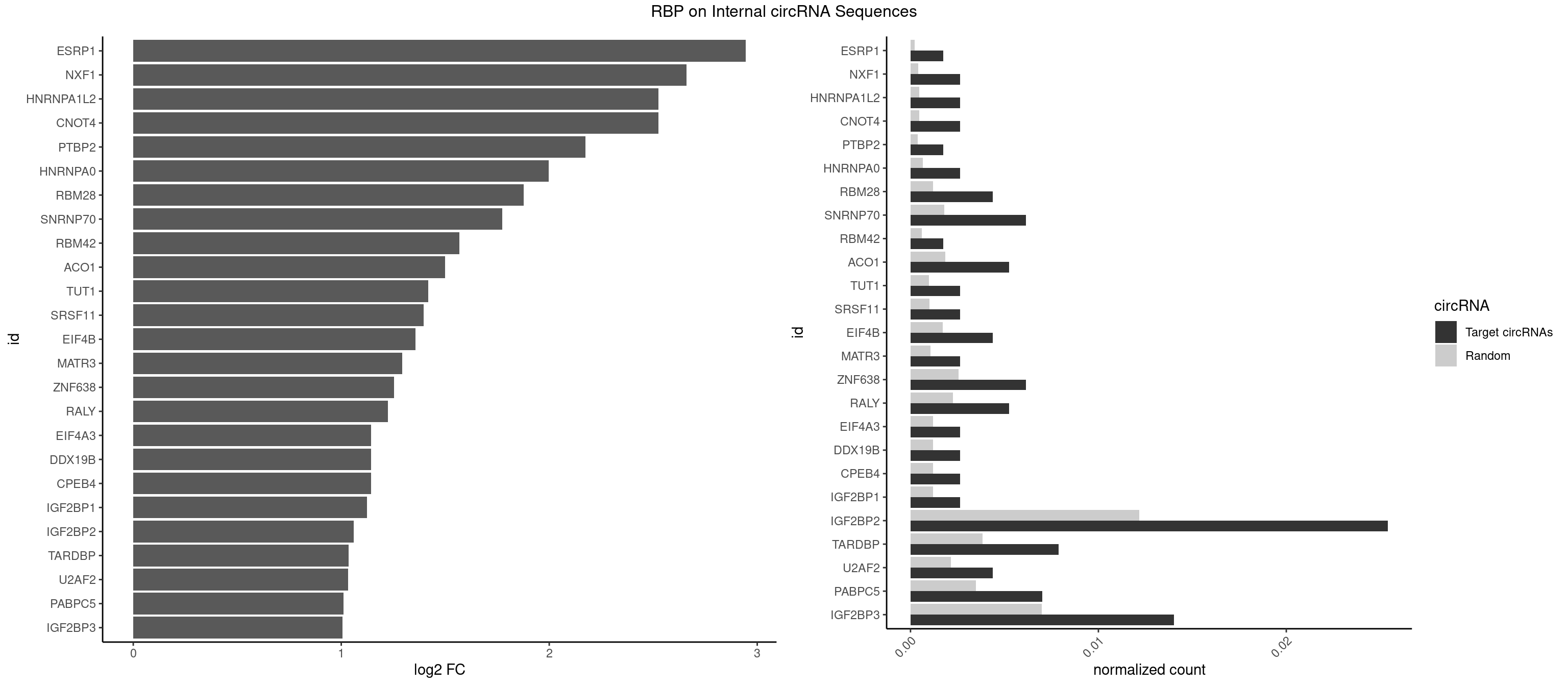
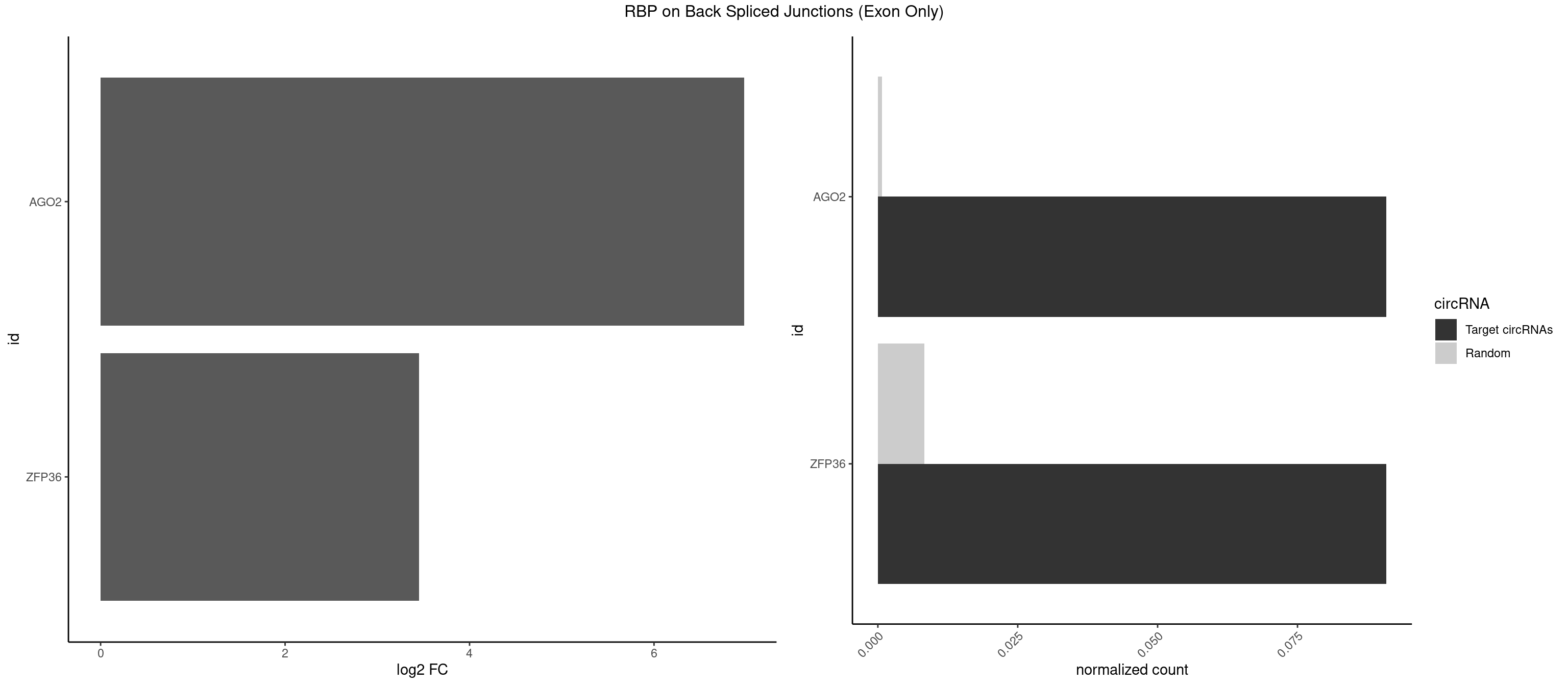
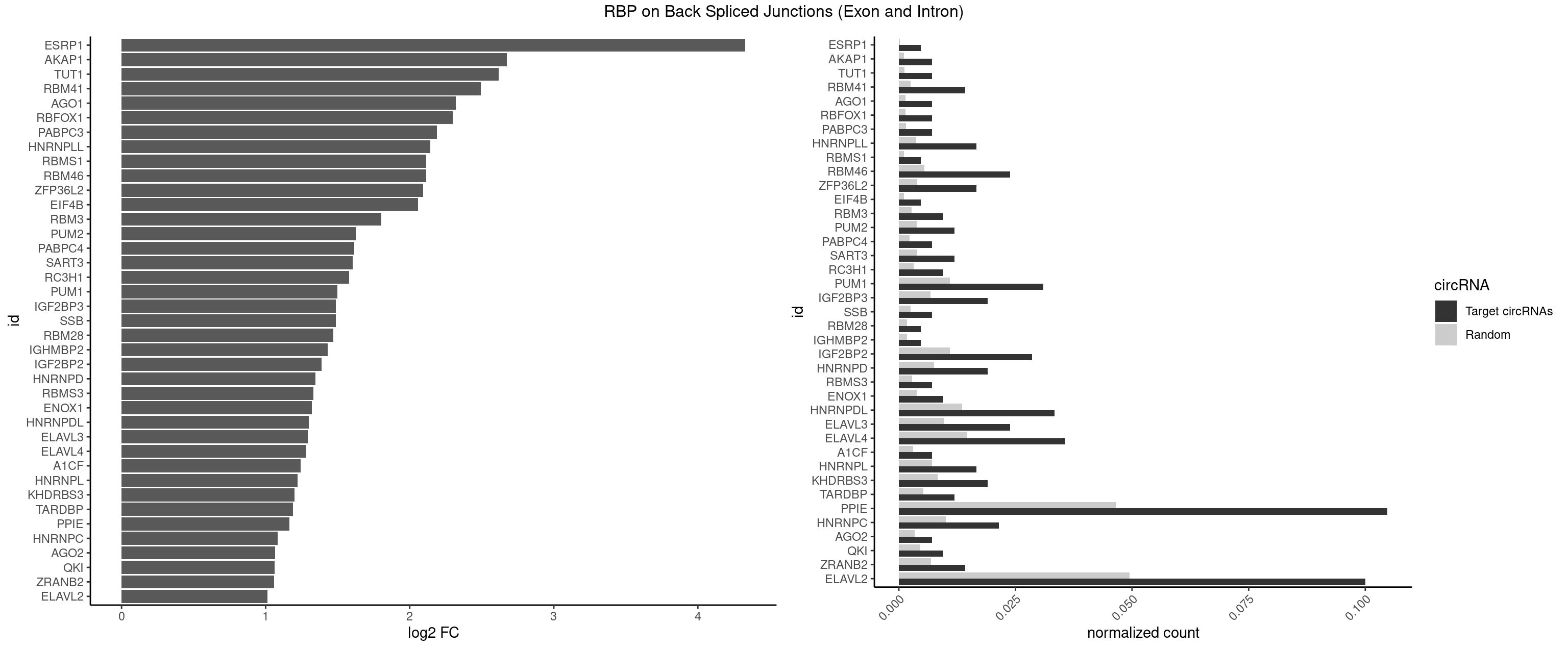
| ESRP1 |
1 |
46 |
0.004761905 |
0.0002367997 |
4.329800 |
AGGGAU |
AGGGAU |
| AKAP1 |
2 |
221 |
0.007142857 |
0.0011185006 |
2.674935 |
AUAUAU,UAUAUA |
AUAUAU,UAUAUA |
| TUT1 |
2 |
230 |
0.007142857 |
0.0011638452 |
2.617602 |
GAUACU |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM41 |
5 |
502 |
0.014285714 |
0.0025342604 |
2.494937 |
AUACUU,UACAUU,UACUUU,UUACUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| AGO1 |
2 |
283 |
0.007142857 |
0.0014308746 |
2.319604 |
AGGUAG,GUAGUA |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 |
2 |
287 |
0.007142857 |
0.0014510278 |
2.299426 |
AGCAUG,GCAUGU |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC3 |
2 |
310 |
0.007142857 |
0.0015669085 |
2.188580 |
AAAAAC,AAAACA |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPLL |
6 |
748 |
0.016666667 |
0.0037736800 |
2.142922 |
ACACAC,ACACCA,CACACA,CACACC,CACUGC,GCACAC |
ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBMS1 |
1 |
217 |
0.004761905 |
0.0010983474 |
2.116204 |
UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBM46 |
9 |
1091 |
0.023809524 |
0.0055018138 |
2.113560 |
AAUGAA,AUCAUU,AUGAAA,AUGAAU |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZFP36L2 |
6 |
774 |
0.016666667 |
0.0039046755 |
2.093691 |
AUUUAU,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| EIF4B |
1 |
226 |
0.004761905 |
0.0011436921 |
2.057840 |
UUGGAC |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM3 |
3 |
541 |
0.009523810 |
0.0027307537 |
1.802240 |
AAACUA,GAUACU |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PUM2 |
4 |
764 |
0.011904762 |
0.0038542926 |
1.627001 |
GUACAU,UAUAUA,UGUAAA,UGUAUA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC4 |
2 |
462 |
0.007142857 |
0.0023327287 |
1.614483 |
AAAAAA,AAAAAG |
AAAAAA,AAAAAG |
| SART3 |
4 |
777 |
0.011904762 |
0.0039197904 |
1.602690 |
AAAAAA,AAAAAC,GAAAAA |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RC3H1 |
3 |
631 |
0.009523810 |
0.0031841999 |
1.580608 |
CCUUCU,CUUCUG,UUCUGU |
CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PUM1 |
12 |
2172 |
0.030952381 |
0.0109482064 |
1.499356 |
AAUGUU,AGAUAA,GUACAU,UAAUGU,UAUAUA,UGUAAA,UGUAUA,UUAAUG,UUUAAU |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| IGF2BP3 |
7 |
1348 |
0.019047619 |
0.0067966546 |
1.486714 |
AAAAAC,AAAACA,AAACUC,AACUCA,AAUUCA,ACACAC,CACACA |
AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| SSB |
2 |
505 |
0.007142857 |
0.0025493753 |
1.486358 |
UGUUUU |
CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBM28 |
1 |
340 |
0.004761905 |
0.0017180572 |
1.470761 |
AGUAGU |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| IGHMBP2 |
1 |
350 |
0.004761905 |
0.0017684401 |
1.429061 |
AAAAAA |
AAAAAA |
| IGF2BP2 |
11 |
2163 |
0.028571429 |
0.0109028617 |
1.389866 |
AAAAAC,AAAACA,AAACUC,AACUCA,AAUUCA,ACACAC,CAAUUC,CACACA,CCAUUC,GAAUAC,GCACAC |
AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| HNRNPD |
7 |
1488 |
0.019047619 |
0.0075020153 |
1.344261 |
AAAAAA,AUUUAU,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| RBMS3 |
2 |
562 |
0.007142857 |
0.0028365578 |
1.332360 |
AUAUAU,UAUAUA |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ENOX1 |
3 |
756 |
0.009523810 |
0.0038139863 |
1.320239 |
AGACAG,AGUACA,UAGACA |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| HNRNPDL |
13 |
2689 |
0.033333333 |
0.0135530028 |
1.298353 |
AAUACC,ACACCA,ACAUUA,ACUAGA,ACUUUA,AUACCA,AUUAGG,CACCAG,CAUUAG,CUAACU,CUAAGU,GGACUA,UAAGUA |
AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| ELAVL3 |
9 |
1928 |
0.023809524 |
0.0097188634 |
1.292679 |
AUUUAU,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ELAVL4 |
14 |
2916 |
0.035714286 |
0.0146966949 |
1.281010 |
AAAAAA,AUUUAU,UAUUUU,UCUAAU,UUAUUU,UUUAUU,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| A1CF |
2 |
598 |
0.007142857 |
0.0030179363 |
1.242939 |
AUAAUU,UUAAUU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| HNRNPL |
6 |
1419 |
0.016666667 |
0.0071543732 |
1.220068 |
AAACAA,ACACAC,ACACCA,CACACA,CACACC,CAUAAA |
AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| KHDRBS3 |
7 |
1646 |
0.019047619 |
0.0082980653 |
1.198764 |
AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,UUAAAC,UUUUUU |
AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| TARDBP |
4 |
1034 |
0.011904762 |
0.0052146312 |
1.190902 |
GAAUGU,UGAAUG,UUGUGC,UUGUUC |
GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| PPIE |
43 |
9262 |
0.104761905 |
0.0466696896 |
1.166556 |
AAAAAA,AAAAAU,AAAAUA,AAAUAU,AUAAAA,AUAAUU,AUAUAU,AUAUUA,AUUAAA,AUUUAU,AUUUUU,UAAAAA,UAAAAU,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUU,UUAAAA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPC |
8 |
2006 |
0.021428571 |
0.0101118501 |
1.083489 |
AUUUUU,CUUUUU,GGAUAC,UUUUUA,UUUUUC,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| AGO2 |
2 |
677 |
0.007142857 |
0.0034159613 |
1.064210 |
AAAAAA,UAAAGU |
AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| QKI |
3 |
904 |
0.009523810 |
0.0045596534 |
1.062615 |
ACACAC,ACUUAU,UCUAAU |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ZRANB2 |
5 |
1360 |
0.014285714 |
0.0068571141 |
1.058900 |
AAAGGU,AGGUAA,AGGUAG,AGGUUA,GUAAAG |
AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ELAVL2 |
41 |
9826 |
0.100000000 |
0.0495112858 |
1.014171 |
AAUUUG,AUACUU,AUUUAU,AUUUUC,AUUUUU,CUUAUA,CUUUUU,GUUUUA,GUUUUU,UAAUUU,UACUUU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUUU,UCAUUU,UCUUAU,UUAAUU,UUACUU,UUAUAU,UUAUUU,UUCUUU,UUUAAU,UUUAGU,UUUAUA,UUUAUU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |