circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000089154:-:12:120134290:120153909
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000089154:-:12:120134290:120153909 | ENSG00000089154 | ENST00000300648 | - | 12 | 120134291 | 120153909 | 3616 | GUGUGGCUUGGCGUUGGCCCUCAACAAGCUCUCCCAGUAUUUGGACAGCUCUCAGGUGAAGCCACUCUUUCAGUUUUUUGUCCCUGAUGCCCUCAAUGACCGACACCCAGAUGUCCGGAAGUGCAUGUUGGAUGCAGCCCUCGCAACGCUCAACACUCAUGGGAAGGAGAACGUCAACUCGCUGUUGCCAGUAUUCGAGGAGUUCCUGAAGAACGCGCCCAAUGAUGCCAGCUAUGAUGCUGUGCGACAGAGUGUGGUGGUCCUGAUGGGCUCUCUGGCCAAGCACCUGGACAAGAGUGACCCCAAAGUGAAGCCCAUUGUUGCCAAGCUCAUCGCUGCCCUCUCCACCCCCUCCCAGCAGGUCCAGGAGUCCGUAGCCAGCUGCUUGCCACCCCUUGUGCCAGCCAUCAAGGAGGAUGCUGGAGGGAUGAUCCAGAGGCUUAUGCAGCAGCUGCUGGAGUCAGACAAGUACGCAGAGCGCAAAGGGGCCGCCUAUGGCCUGGCGGGCCUGGUGAAGGGCCUGGGCAUCCUCUCGCUGAAGCAACAGGAGAUGAUGGCGGCACUGACUGAUGCCAUCCAAGAUAAGAAGAACUUCCGCCGGCGAGAGGGAGCCCUCUUUGCCUUCGAGAUGCUCUGCACCAUGCUGGGGAAACUUUUUGAGCCGUAUGUGGUUCACGUGCUGCCCCAUCUGCUCCUGUGCUUUGGGGAUGGAAACCAGUAUGUGCGUGAGGCUGCAGAUGACUGUGCCAAGGCUGUGAUGAGCAACUUGAGUGCUCACGGGGUGAAGCUGGUGCUCCCCUCCUUACUGGCUGCCCUGGAGGAGGAAUCGUGGCGGACCAAAGCUGGGUCAGUGGAGCUUCUUGGGGCAAUGGCGUACUGUGCUCCUAAGCAGCUGUCAUCCUGUCUACCCAACAUUGUGCCCAAGCUUACGGAGGUGCUGACCGACUCCCAUGUCAAAGUCCAGAAGGCUGGACAGCAGGCGCUCAGGCAGAUCGGCUCCGUUAUCAGGAACCCGGAGAUCCUGGCCAUUGCUCCAGUCCUCCUGGAUGCCCUGACGGAUCCCUCCAGGAAGACCCAGAAGUGCUUGCAGACCCUGCUGGACACCAAGUUUGUCCACUUCAUUGAUGCCCCAUCCCUGGCCCUCAUCAUGCCCAUUGUCCAGAGAGCCUUCCAGGACCGUUCCACGGACACGCGGAAGAUGGCAGCCCAGAUUAUUGGCAACAUGUACUCCCUGACAGACCAGAAGGACUUGGCUCCGUACCUGCCCAGCGUGACGCCUGGCCUGAAAGCAUCGCUUUUGGACCCUGUGCCUGAGGUGCGGACCGUAUCUGCAAAGGCCCUUGGGGCCAUGGUGAAGGGCAUGGGGGAGUCGUGCUUUGAGGACUUGCUGCCGUGGCUGAUGGAGACACUGACCUAUGAGCAGAGCUCUGUGGAUCGCUCAGGCGCUGCACAGGGGUUGGCUGAGGUCAUGGCCGGUUUGGGGGUGGAGAAGUUGGAGAAGUUGAUGCCAGAAAUCGUGGCUACAGCCAGCAAAGUGGACAUUGCACCCCAUGUCCGAGAUGGCUACAUUAUGAUGUUUAACUACCUGCCCAUCACCUUUGGAGACAAGUUUACUCCUUAUGUGGGGCCCAUCAUCCCCUGUAUCCUCAAAGCUCUUGCUGAUGAGAAUGAGUUUGUGCGUGACACCGCCCUGCGCGCGGGCCAGCGGGUUAUCUCCAUGUACGCUGAGACAGCCAUCGCCCUGCUGCUGCCCCAGCUAGAGCAAGGCCUCUUUGAUGACCUUUGGAGAAUCAGGUUCAGCUCUGUUCAGCUCCUUGGGGAUCUCCUGUUUCACAUCUCAGGAGUCACUGGGAAGAUGACCACAGAAACUGCCUCUGAGGAUGAUAACUUUGGAACUGCCCAGUCCAACAAGGCGAUCAUCACUGCCCUGGGGGUAGAGCGGCGGAACCGGGUGUUGGCAGGGCUGUACAUGGGCCGCUCAGACACCCAGCUGGUGGUGCGGCAGGCGUCCCUGCAUGUCUGGAAGAUUGUUGUCUCCAAUACCCCCCGCACCUUGCGUGAGAUCCUACCCACUCUCUUUGGGCUCCUGCUGGGUUUCCUGGCCAGCACGUGUGCAGAUAAGAGAACGAUUGCAGCGAGAACAUUGGGAGAUCUUGUGCGGAAGUUAGGGGAGAAAAUCCUCCCCGAGAUCAUCCCCAUCCUUGAGGAAGGCCUGAGGUCUCAGAAGAGCGAUGAGAGGCAGGGUGUGUGCAUUGGCCUAAGUGAGAUCAUGAAGUCCACCAGCCGGGAUGCCGUGCUGUAUUUCUCUGAAUCCCUCGUGCCCACGGCAAGGAAGGCUUUGUGUGACCCACUGGAGGAGGUCAGAGAGGCGGCAGCCAAGACUUUCGAGCAGCUGCAUUCCACCAUCGGCCACCAGGCUCUGGAGGACAUUCUCCCAUUUUUACUAAAGCAGCUGGAUGACGAGGAGGUGUCAGAGUUUGCCUUGGAUGGUCUGAAGCAAGUCAUGGCUAUUAAGAGUCGUGUGGUGCUGCCCUACCUUGUGCCCAAGCUGACAACGCCACCUGUCAACACCCGGGUGCUGGCUUUCCUUUCGUCAGUGGCUGGUGAUGCCCUCACCCGUCAUCUUGGCGUGAUCCUCCCAGCGGUCAUGCUGGCCCUGAAGGAAAAGCUUGGGACCCCAGAUGAGCAGCUGGAGAUGGCCAAUUGUCAGGCUGUGAUCCUCUCCGUAGAGGAUGACACAGGGCACCGGAUCAUCAUCGAGGAUCUGCUGGAGGCCACCCGCAGCCCUGAGGUGGGCAUGAGGCAAGCUGCUGCCAUCAUCCUCAACAUCUACUGUUCCCGCUCAAAGGCUGACUACACCAGCCACCUGCGGAGCCUGGUCUCGGGCCUGAUCCGCCUCUUCAAUGACUCCAGCCCUGUGGUUCUGGAGGAGAGCUGGGAUGCCCUAAAUGCCAUCACUAAGAAGCUGGAUGCUGGCAACCAGUUGGCACUCAUUGAAGAGCUGCACAAGGAAAUCCGGCUCAUAGGGAACGAGAGCAAAGGCGAGCAUGUGCCAGGAUUCUGCCUCCCGAAGAAGGGAGUGACCUCCAUCCUUCCAGUGUUGCGGGAAGGAGUCCUGACUGGCAGCCCUGAGCAGAAGGAGGAGGCAGCCAAAGCCUUAGGCUUGGUAAUCCGCCUGACCUCGGCUGACGCCCUGAGGCCCUCCGUGGUCAGCAUCACUGGCCCUCUGAUCCGCAUCCUGGGGGACAGGUUCAGCUGGAAUGUGAAGGCGGCUCUGCUCGAGACACUCAGCCUCUUGUUGGCUAAGGUUGGGAUUGCCCUGAAGCCCUUCCUGCCCCAGCUGCAGACCACUUUCACCAAAGCCCUGCAGGACUCCAACCGGGGGGUGCGCCUGAAGGCCGCAGAUGCUCUGGGGAAGCUCAUUUCCAUCCACAUUAAGGUGGACCCCCUCUUCACAGAGCUGCUCAAUGGCAUCCGCGCCAUGGAGGACCCAGGUGUCAGGGACACCAUGCUGCAGGCCCUGAGGUUUGUGAUUCAGGGAGCAGGGGCCAAAGUGGAUGCCGUCAUCCGGAAAAACAUCGUCUCACUCCUGCUGAGCAUGCUGGGACACGAUGAGGUGUGGCUUGGCGUUGGCCCUCAACAAGCUCUCCCAGUAUUUGGACAGCU | circ |
| ENSG00000089154:-:12:120134290:120153909 | ENSG00000089154 | ENST00000300648 | - | 12 | 120134291 | 120153909 | 22 | GACACGAUGAGGUGUGGCUUGG | bsj |
| ENSG00000089154:-:12:120134290:120153909 | ENSG00000089154 | ENST00000300648 | - | 12 | 120153900 | 120154109 | 210 | AUGGCAAGUCUCCAUAUGACAAGUGGGUUGUUCCAGAACAUUGGGUGAGCACUGGGUGGUGGGCUUCAGGCUGUUCUCUGCCCCCAAAACAGUGUUUGCCUCAGGCAAGCUCCCUCUCCUUGCCGUGCACCAGCAGAGGUUCCACUGGCAGAGGAGGCCCCCUGACUGCCCCUCCUGAUGUGCUCCCACUUCUUUCCCAGGUGUGGCUUG | ie_up |
| ENSG00000089154:-:12:120134290:120153909 | ENSG00000089154 | ENST00000300648 | - | 12 | 120134091 | 120134300 | 210 | ACACGAUGAGGUACGGCUGCAGGCAGGACUAGUGGCAGCACUGGACCACUGGUUUCCCUCCUCCUCCUUAGGUUGAGUUCUUCACUUUGUGUUAGAACCAGUACAGCUCAGUAGUAAUAAGCAUUCCUGACAUUUCUAUCACCCUGUAUGCUUCACCAACAUUUCCUCGGGUUUCAAGGGAAACGGGGCAGGUAUUGUGGAUAUAUUGGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
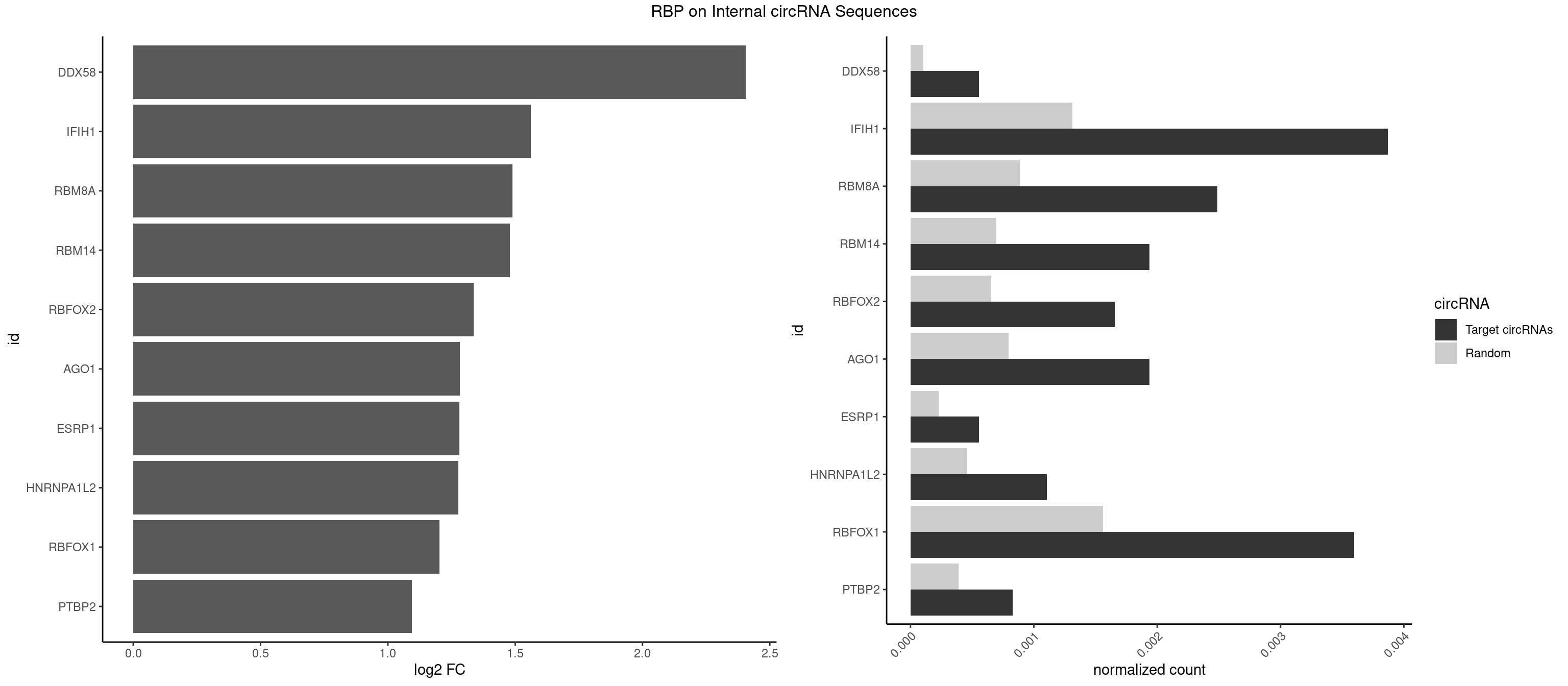
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| DDX58 | 1 | 71 | 0.0005530973 | 0.0001043427 | 2.406204 | GCGCGC | GCGCGC |
| IFIH1 | 13 | 904 | 0.0038716814 | 0.0013115296 | 1.561710 | CGCGGA,GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBM8A | 8 | 611 | 0.0024889381 | 0.0008869128 | 1.488666 | ACGCGC,CGCGCC,CGCGCG,GCGCGC,GUGCGC,UGCGCC,UGCGCG | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBM14 | 6 | 478 | 0.0019358407 | 0.0006941687 | 1.479602 | CGCGCC,CGCGCG,CGCGGG,GCGCGC,GCGCGG | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBFOX2 | 5 | 452 | 0.0016592920 | 0.0006564894 | 1.337724 | UGACUG,UGCAUG | UGACUG,UGCAUG |
| AGO1 | 6 | 548 | 0.0019358407 | 0.0007956130 | 1.282822 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| ESRP1 | 1 | 156 | 0.0005530973 | 0.0002275250 | 1.281508 | AGGGAU | AGGGAU |
| HNRNPA1L2 | 3 | 314 | 0.0011061947 | 0.0004564992 | 1.276921 | AUAGGG,UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBFOX1 | 12 | 1077 | 0.0035951327 | 0.0015622419 | 1.202427 | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PTBP2 | 2 | 267 | 0.0008296460 | 0.0003883867 | 1.095002 | CUCUCU | CUCUCU |
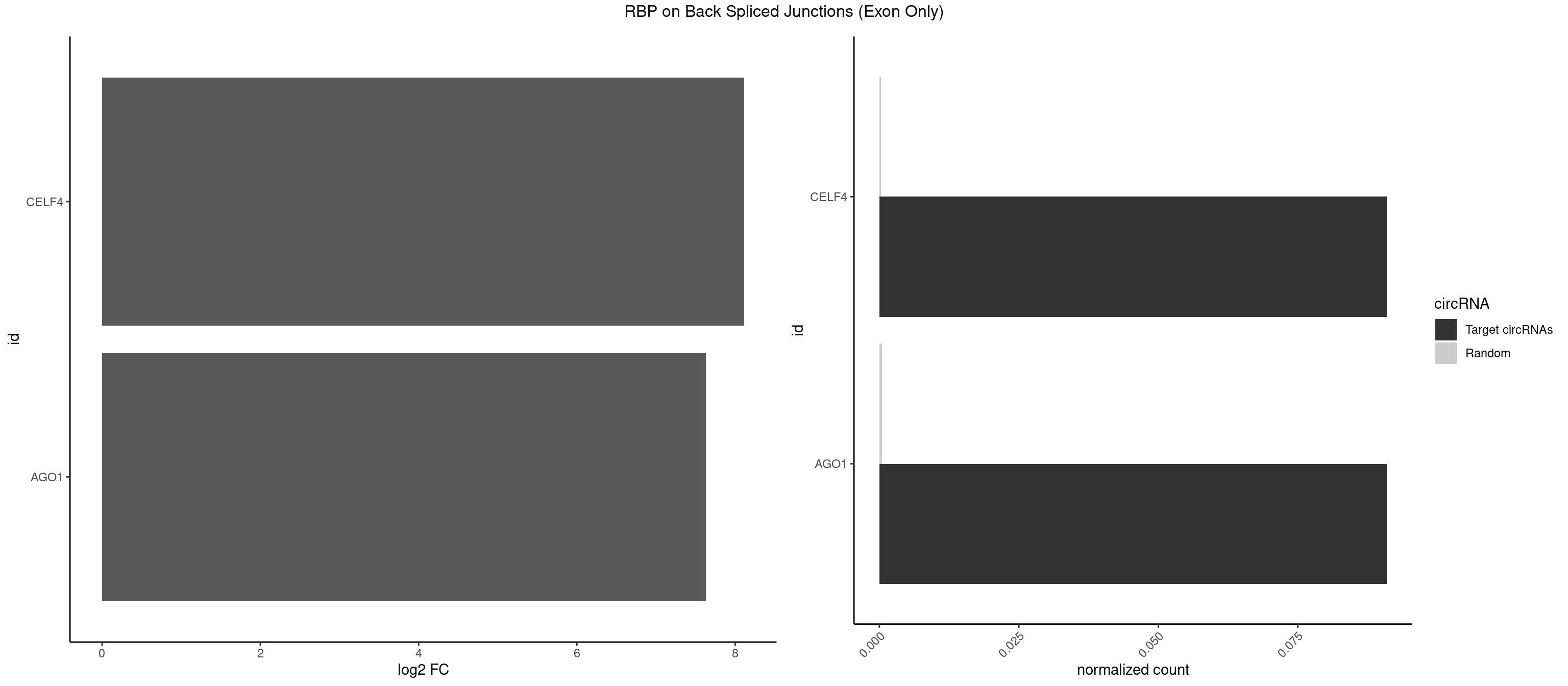
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CELF4 | 1 | 4 | 0.09090909 | 0.0003274823 | 8.116864 | GGUGUG | GGUGUG,GUGUUG,UGUGUG |
| AGO1 | 1 | 6 | 0.09090909 | 0.0004584752 | 7.631437 | UGAGGU | AGGUAG,GAGGUA,UGAGGU |
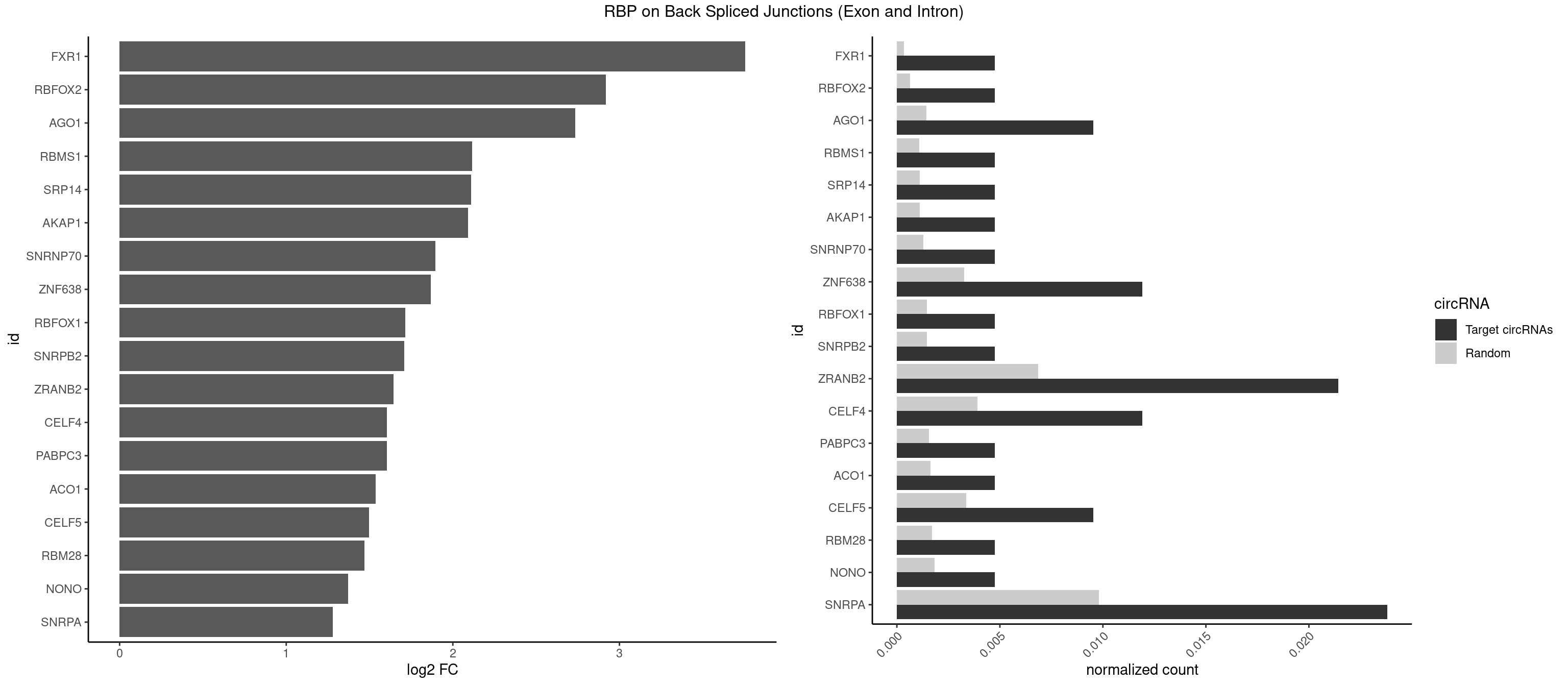
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| AGO1 | 3 | 283 | 0.009523810 | 0.0014308746 | 2.734641 | GAGGUA,GUAGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CCUGUA | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | GGUUGU,GUUCUU,GUUGUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | GUAUUG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ZRANB2 | 8 | 1360 | 0.021428571 | 0.0068571141 | 1.643862 | AGGGAA,AGGUAC,AGGUAU,GAGGUU,GGGUGA,GGUGGU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GGUGUG,GUGUGG,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGG,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SNRPA | 9 | 1947 | 0.023809524 | 0.0098145909 | 1.278539 | AGUAGU,AUUCCU,CAGUAG,GUAUGC,GUUUCC,UAUGCU,UGCACC,UUCCUG,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.