circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000143748:-:1:224303722:224305166
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000143748:-:1:224303722:224305166 | ENSG00000143748 | MSTRG.3125.12 | - | 1 | 224303723 | 224305166 | 345 | GAUUCAAAAGAUUCUUCUCUUUUGGAGAGUGAUAUGAAACGGAAAGGCAAGCUAAAGAAUAAAGGAAGCAAAAGGAAGAAAGAAGAUCUUCAGGAAGUAGAUGGAGAAAUUGAAGCUGUCCUGCAAAAGAAAGCUAAAGCCAGGGGGUUAGAAUUCCAGAUCUCCAACGUGAAGUUUGAAGAUGUGGGAGGCAAUGAUAUGACAUUAAAAGAGGUCUGCAAGAUGCUCAUACACAUGCGUCACCCGGAGGUGUACCACCACCUGGGCGUCGUGCCCCCUCGUGGAGUUCUCCUUCAUGGACCACCAGGCUGUGGGAAGACAUUACUUGCACAUGCAAUUGCUGGGGAUUCAAAAGAUUCUUCUCUUUUGGAGAGUGAUAUGAAACGGAAAGGCAA | circ |
| ENSG00000143748:-:1:224303722:224305166 | ENSG00000143748 | MSTRG.3125.12 | - | 1 | 224303723 | 224305166 | 22 | CAAUUGCUGGGGAUUCAAAAGA | bsj |
| ENSG00000143748:-:1:224303722:224305166 | ENSG00000143748 | MSTRG.3125.12 | - | 1 | 224305157 | 224305366 | 210 | UUAAACUGAGCUUUAAGGGAAUGAGGAGAAGGACUUUCACAAUAGACAGCAUCUUUGAGCAAAGAGAUUGGGAAUAGUUAACAGAGGUGAAAGUGCUAGGGCAGGAAUGAAUUUUCUUUACAAUGCCCCUCUUUAAAAAGUAAAACAAUUAUUGGCAUAGAAUUUUAAGCAUGGCAUAUUUAAAUUUAUUUUCUUUCCAGGAUUCAAAAG | ie_up |
| ENSG00000143748:-:1:224303722:224305166 | ENSG00000143748 | MSTRG.3125.12 | - | 1 | 224303523 | 224303732 | 210 | AAUUGCUGGGGUGAGGCUUGCUGACAUUUGUUUGCUUUUGCUGUUUUUGUUGUUAUUAUUUGUUUUUGUUUUUUCUCUUUGGUCAACUUAAACCAGACAUGGCAUUCUUCAGGAAGAGAACAUAGUCAUUGGAACUGAUCCCUUUGUUAUCAUGCUGGUAUUAUUUGUGGAAUUUAUUCAUUUAGUUUAUUUAUUGUUUCUCACGAGCAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
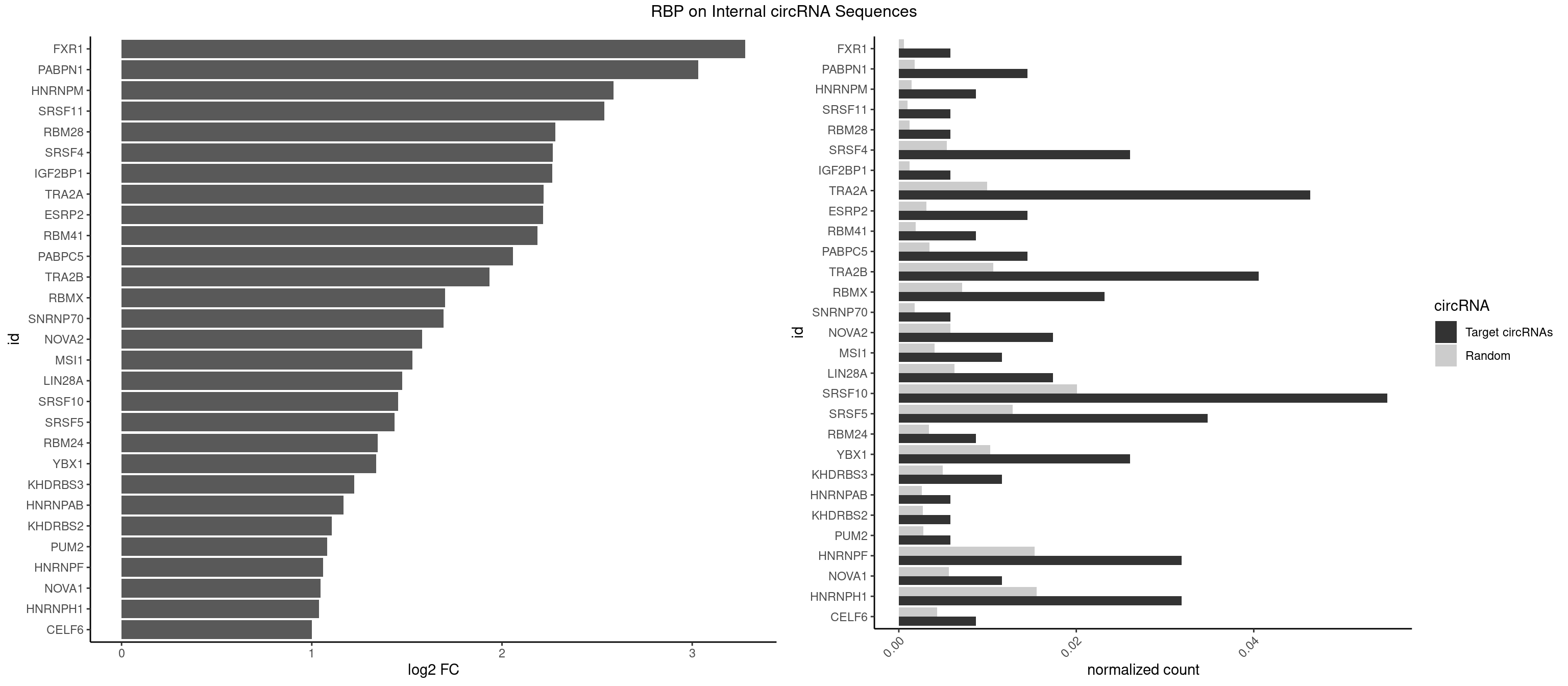
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 411 | 0.005797101 | 0.0005970720 | 3.279355 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| PABPN1 | 4 | 1222 | 0.014492754 | 0.0017723764 | 3.031575 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| HNRNPM | 2 | 999 | 0.008695652 | 0.0014492040 | 2.585034 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| SRSF11 | 1 | 688 | 0.005797101 | 0.0009985015 | 2.537495 | AAGAAG | AAGAAG |
| RBM28 | 1 | 822 | 0.005797101 | 0.0011926949 | 2.281107 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SRSF4 | 8 | 3740 | 0.026086957 | 0.0054214720 | 2.266572 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| IGF2BP1 | 1 | 831 | 0.005797101 | 0.0012057377 | 2.265416 | CACCCG | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| TRA2A | 15 | 6871 | 0.046376812 | 0.0099589296 | 2.219341 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGGAAG,GAAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ESRP2 | 4 | 2150 | 0.014492754 | 0.0031172377 | 2.216992 | GGGAAG,GGGGAU,UGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| RBM41 | 2 | 1318 | 0.008695652 | 0.0019115000 | 2.185589 | UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PABPC5 | 4 | 2400 | 0.014492754 | 0.0034795387 | 2.058364 | AGAAAG,AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TRA2B | 13 | 7329 | 0.040579710 | 0.0106226650 | 1.933613 | AAAGAA,AAGAAG,AAGAAU,AAGGAA,AGAAGA,AGGAAG,GAAAGA,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBMX | 7 | 4925 | 0.023188406 | 0.0071387787 | 1.699654 | AAGAAG,AAGGAA,AGGAAG,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SNRNP70 | 1 | 1237 | 0.005797101 | 0.0017941145 | 1.692060 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| NOVA2 | 5 | 4013 | 0.017391304 | 0.0058171047 | 1.579993 | ACCACC,AGACAU,GAGGCA | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| MSI1 | 3 | 2770 | 0.011594203 | 0.0040157442 | 1.529664 | AGGAAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| LIN28A | 5 | 4315 | 0.017391304 | 0.0062547643 | 1.475339 | CGGAGG,GGAGAA,UGGAGA,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SRSF10 | 18 | 13860 | 0.055072464 | 0.0200874160 | 1.455039 | AAAAGA,AAAGAA,AAAGAG,AAAGGA,AAGAAA,AAGAAG,AAGACA,AAGAGG,AAGGAA,GAAAGA,GAGAAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| SRSF5 | 11 | 8869 | 0.034782609 | 0.0128544391 | 1.436099 | AAGAAG,AGAAGA,AGGAAG,AUAAAG,GAAGAA,GGAAGA,UAAAGG,UGCGUC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| RBM24 | 2 | 2357 | 0.008695652 | 0.0034172229 | 1.347470 | AGAGUG,GAGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| YBX1 | 8 | 7119 | 0.026086957 | 0.0103183321 | 1.338119 | ACCACC,CACCAC,CCACCA,GGUCUG,GUCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| KHDRBS3 | 3 | 3429 | 0.011594203 | 0.0049707696 | 1.221863 | AAUAAA,AUAAAG,AUUAAA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPAB | 1 | 1782 | 0.005797101 | 0.0025839306 | 1.165764 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| KHDRBS2 | 1 | 1858 | 0.005797101 | 0.0026940701 | 1.105544 | AAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| PUM2 | 1 | 1890 | 0.005797101 | 0.0027404447 | 1.080922 | GUAGAU | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPF | 10 | 10561 | 0.031884058 | 0.0153064921 | 1.058692 | AGGAAG,AUGUGG,CUGGGG,GGGAAG,GGGAGG,UGGGAA,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| NOVA1 | 3 | 3872 | 0.011594203 | 0.0056127669 | 1.046620 | ACCACC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| HNRNPH1 | 10 | 10717 | 0.031884058 | 0.0155325680 | 1.037539 | AGGAAG,AUGUGG,CUGGGG,GGGAAG,GGGAGG,GGGCGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| CELF6 | 2 | 2997 | 0.008695652 | 0.0043447134 | 1.001033 | UGUGGG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
RBP on BSJ (Exon Only)
Plot
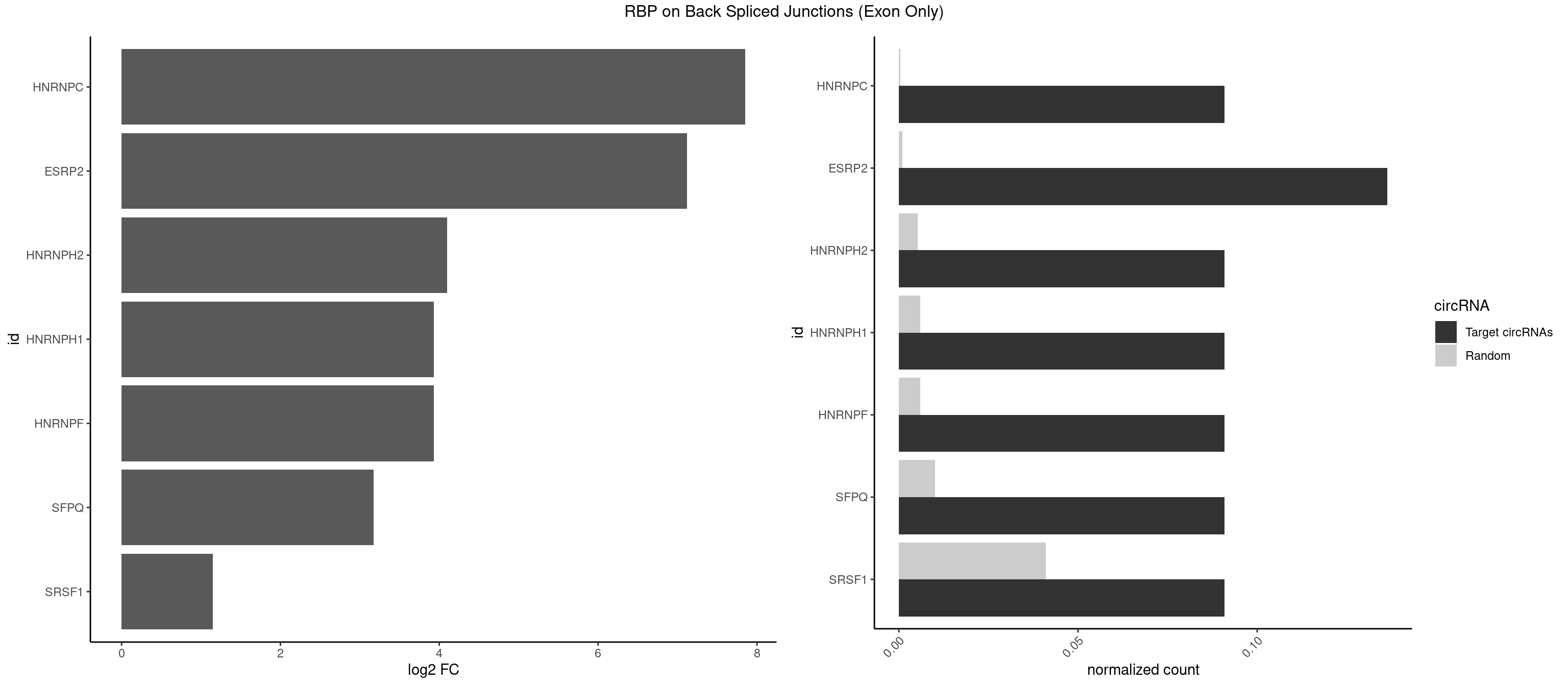
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPC | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GGAUUC | GGAUAU,GGAUUC,GGGUAC |
| ESRP2 | 2 | 14 | 0.13636364 | 0.0009824469 | 7.116864 | GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,UGGGAA,UGGGGA |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.0053052135 | 4.098942 | CUGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPF | 1 | 90 | 0.09090909 | 0.0059601782 | 3.930997 | CUGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.0059601782 | 3.930997 | CUGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | GGGGAU | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF1 | 1 | 625 | 0.09090909 | 0.0410007860 | 1.148773 | GGAUUC | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
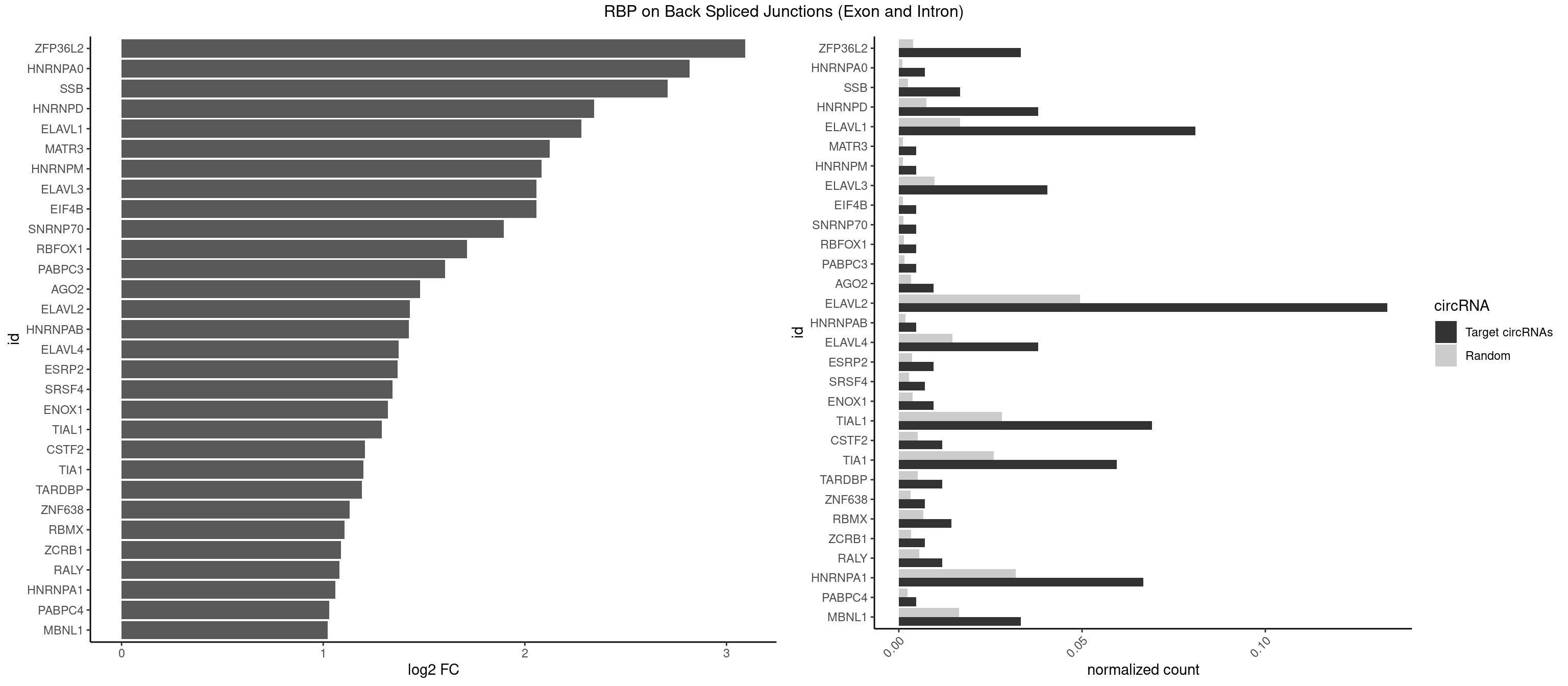
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZFP36L2 | 13 | 774 | 0.033333333 | 0.003904676 | 3.093691 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.001012696 | 2.818299 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| SSB | 6 | 505 | 0.016666667 | 0.002549375 | 2.708750 | CUGUUU,GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPD | 15 | 1488 | 0.038095238 | 0.007502015 | 2.344261 | AAUUUA,AUUUAU,UAUUUA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ELAVL1 | 33 | 3309 | 0.080952381 | 0.016676743 | 2.279236 | AUUUAU,UAGUUU,UAUUUA,UAUUUU,UGUUUU,UUAGUU,UUAUUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.001093309 | 2.122837 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.001123539 | 2.083489 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ELAVL3 | 16 | 1928 | 0.040476190 | 0.009718863 | 2.058214 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.001143692 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.001279726 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.001451028 | 1.714464 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.001566909 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| AGO2 | 3 | 677 | 0.009523810 | 0.003415961 | 1.479247 | AAAGUG,AAGUGC,AGUGCU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| ELAVL2 | 55 | 9826 | 0.133333333 | 0.049511286 | 1.429208 | AAUUUA,AAUUUU,AUAUUU,AUUAUU,AUUUAA,AUUUAG,AUUUAU,AUUUUA,AUUUUC,CUUUAA,GUUUUU,UAGUUA,UAUUAU,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UUAGUU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUUAAG,UUUAGU,UUUAUU,UUUCUU,UUUUAA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.001773478 | 1.424957 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| ELAVL4 | 15 | 2916 | 0.038095238 | 0.014696695 | 1.374119 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.003688029 | 1.368689 | GGGAAU,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SRSF4 | 2 | 558 | 0.007142857 | 0.002816405 | 1.342647 | AGGAAG,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ENOX1 | 3 | 756 | 0.009523810 | 0.003813986 | 1.320239 | AGACAG,CAGACA,UAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| TIAL1 | 28 | 5597 | 0.069047619 | 0.028204353 | 1.291674 | AAAUUU,AAUUUU,AUUUUA,AUUUUC,CUUUUG,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUUAAA,UUUAUU,UUUUAA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.005154172 | 1.207726 | UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TIA1 | 24 | 5140 | 0.059523810 | 0.025901854 | 1.200411 | AUUUUC,CUUUUG,GUUUUU,UAUUUU,UUAUUU,UUUAUU,UUUUCU,UUUUGU,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.005214631 | 1.190902 | GAAUGA,GUUGUU,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZNF638 | 2 | 646 | 0.007142857 | 0.003259774 | 1.131729 | GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBMX | 5 | 1316 | 0.014285714 | 0.006635429 | 1.106311 | AAGUAA,AGAAGG,AGGAAG,GAAGGA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.003360540 | 1.087808 | ACUUAA,AUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RALY | 4 | 1117 | 0.011904762 | 0.005632809 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPA1 | 27 | 6340 | 0.066666667 | 0.031947803 | 1.061249 | AAAGAG,AAUUUA,AGGAAG,AGGGCA,AUAGAA,AUUUAA,AUUUAU,CAAAGA,GAGGAG,GGACUU,GGCAGG,GGGCAG,UAGACA,UAGGGC,UAGUUA,UUAUUU,UUUAUU,UUUUUU | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.002332729 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
| MBNL1 | 13 | 3258 | 0.033333333 | 0.016419790 | 1.021530 | AUGCCC,CUUGCU,GCUUGC,GCUUUU,GUGCUA,UGCUGU,UGCUUU,UUGCUG,UUGCUU,UUUGCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.