circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000114316:-:3:49276426:49327816
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000114316:-:3:49276426:49327816 | ENSG00000114316 | MSTRG.22947.1 | - | 3 | 49276427 | 49327816 | 3151 | AUCCUGAGAGUCAGACCUUGAAAGAACACUUAAUUGAUGAAUUGGACUAUGUAUUGGUCCCUACCGAGGCGUGGAAUAAACUACUAAACUGGUACGGCUGUGUAGAAGGCCAGCAACCCAUCGUCAGAAAAGUUGUGGAGCAUGGCCUGUUUGUCAAGCACUGCAAAGUCGAGGUGUAUUUGCUGGAACUGAAGCUCUGUGAGAACAGUGACCCCACCAAUGUGCUGAGUUGCCAUUUCAGCAAGGCAGACACCAUUGCAACCAUCGAGAAAGAGAUGCGGAAGCUAUUCAACAUCCCUGCGGAGCGUGAAACACGGCUCUGGAACAAAUACAUGAGCAACACCUACGAGCAGUUGAGCAAGCUAGACAACACUGUCCAGGAUGCUGGGCUAUACCAGGGUCAGGUGCUAGUAAUUGAGCCUCAAAAUGAAGAUGGCACAUGGCCCAGGCAGACCUUGCAGUCAAAUGGAUCUGGCUUUUCUGCUUCGUAUAAUUGUCAGGAGCCACCAUCCUCUCAUAUACAACCUGGGCUCUGUGGACUUGGAAACCUGGGAAACACCUGCUUCAUGAACUCCGCUUUGCAGUGUUUGAGCAACACUGCACCACUGACUGACUACUUUCUCAAAGAUGAGUAUGAAGCCGAAAUCAACAGAGACAACCCUCUGGGGAUGAAAGGGGAAAUUGCAGAAGCCUAUGCUGAACUCAUUAAGCAGAUGUGGUCUGGAAGGGACGCCCAUGUGGCACCUCGCAUGUUCAAAACUCAAGUAGGACGUUUUGCUCCUCAAUUUUCUGGCUACCAGCAACAAGAUUCUCAGGAGCUGCUGGCCUUUCUUCUAGAUGGAUUGCAUGAAGAUCUGAACCGGGUAAAGAAAAAGCCCUACUUGGAGCUGAAGGAUGCCAAUGGGCGGCCAGAUGCGGUGGUGGCAAAGGAAGCCUGGGAGAAUCACAGGUUGAGGAAUGAUUCUGUGAUUGUGGAUACUUUCCAUGGCCUCUUCAAAUCUACUUUGGUUUGCCCAGAAUGUGCUAAGGUUUCUGUGACCUUUGACCCAUUUUGCUAUCUAACGCUGCCACUGCCCUUGAAGAAAGAUCGAGUUAUGGAGGUUUUCCUGGUUCCUGCUGACCCUCACUGCAGACCUACUCAGUACCGUGUGACUGUGCCGCUGAUGGGGGCUGUGUCCGACCUGUGCGAGGCUCUCUCCAGGCUGUCUGGCAUUGCUGCAGAAAAUAUGGUGGUCGCAGAUGUGUAUAAUCACCGAUUCCACAAAAUUUUCCAAAUGGAUGAAGGUUUAAACCACAUCAUGCCUCGGGAUGACAUUUUCGUGUACGAGGUCUGCAGCACUUCCGUGGAUGGCUCGGAAUGUGUCACGCUUCCAGUCUACUUCAGGGAGAGGAAGUCCAGGCCAUCAAGCACUUCCUCCGCAUCAGCGCUAUAUGGGCAGCCACUAUUGCUUUCUGUCCCCAAGCACAAGUUAACCCUUGAGUCUUUGUACCAGGCUGUUUGUGAUCGUAUCAGCCGCUAUGUGAAACAGCCUUUACCUGAUGAGUUUGGCAGCUCACCCUUGGAGCCAGGGGCCUGCAAUGGCUCCAGGAACAGCUGUGAAGGAGAAGAUGAGGAAGAAAUGGAGCAUCAGGAAGAAGGCAAAGAGCAGCUUUCAGAAACAGAAGGCAGUGGGGAAGAUGAGCCAGGAAAUGACCCCAGUGAGACCACCCAAAAGAAGAUCAAAGGCCAGCCCUGCCCAAAAAGGCUUUUUACCUUCAGUCUUGUGAACUCCUAUGGAACAGCUGACAUAAAUUCACUUGCAGCUGAUGGAAAACUACUUAAACUCAACUCUCGAUCUACACUGGCCAUGGAUUGGGACAGUGAAACUCGGAGACUUUACUAUGAUGAGCAAGAAUCUGAGGCCUACGAGAAGCAUGUGAGCAUGUUGCAGCCUCAGAAGAAGAAGAAGACCACAGUGGCCCUGAGAGACUGCAUCGAGCUCUUCACCACCAUGGAGACCCUUGGGGAGCAUGACCCCUGGUACUGUCCCAACUGUAAGAAGCAUCAACAGGCCACAAAAAAGUUUGACCUAUGGUCCUUGCCCAAGAUCCUGGUGGUCCACCUCAAACGUUUCUCCUACAACAGAUACUGGAGGGAUAAGCUCGACACAGUCGUAGAAUUCCCAAUCAGAGGGCUGAACAUGUCCGAGUUUGUCUGUAACCUGUCAGCAAGGCCUUAUGUGUACGACCUCAUUGCCGUGUCCAAUCAUUAUGGAGCCAUGGGGGUUGGCCACUACACUGCAUAUGCGAAGAACAAACUGAAUGGUAAAUGGUAUUACUUUGAUGAUAGCAACGUGUCCCUGGCCUCUGAGGAUCAGAUAGUGACUAAAGCAGCUUAUGUGCUAUUUUACCAACGUCGAGAUGAUGAAUUUUAUAAGACACCUUCACUUAGCAGUUCUGGUUCCUCUGAUGGAGGGACACGACCAAGCAGCUCUCAGCAGGGCUUUGGGGAUGAUGAGGCUUGCAGCAUGGACACCAACUAAUGCUGACUCCACGAUCCUGCCACCCUGUAGCGCCAGUGUAAUCCCCCAGGAGAACAUCUUUGACACUCUGCAGACUGCUAGUGUUCUGUCUAAAAACCAGACAAGGAAAUACCCUUCUUUUAUGAGCAGAAGGAAACAAAAAAAAAAAAAGAAGACCGUUUACCUAGAAGAAGCUAUGUCAAGAGGCUGAAUUAUUUUUAUUUUUAAACAGGUGGAGAAAUGUCUGAAAAACUAAGAAGAUGCAGAAAGGAACUGACUGCAGCCAUUGACCGGGCCUUUGAAGGAGUUAGUUAUUCCCAGGAGUGCACAGGCCAGCAGAGGCUGGAACUGAGCGCCGCGCCGCUCUCCUUCUCGCUGCCCGUGCACAGGCUCCUCUGCAGAAGACAUCCUCUGGCAGCCUGCUCUUCUGCUGCUCCUUUUGCUGCUGUCCCAUGUGCUCCUGAGAAUGAGAACCCUGCCUUUGCAACAAACCAUGCCCCGGUAAAUGCAAAACCACAUGCUCUGUGCCCCGAGAGAAAACCUCUAACCAGCAAGGAAAAUGUAUUGAUGCAUUCCUCCAUUUUGGCACCUGAAAGAGAGUCUUGGAGAACUGCAGGAGAGGGGGAAAACUGGAGAAAAGAAAAUUUAAGAUCCUGAGAGUCAGACCUUGAAAGAACACUUAAUUGAUGAAUUGGACUAU | circ |
| ENSG00000114316:-:3:49276426:49327816 | ENSG00000114316 | MSTRG.22947.1 | - | 3 | 49276427 | 49327816 | 22 | GAAAAUUUAAGAUCCUGAGAGU | bsj |
| ENSG00000114316:-:3:49276426:49327816 | ENSG00000114316 | MSTRG.22947.1 | - | 3 | 49327807 | 49328016 | 210 | AAAAUCUUCAUGGAUAAAGAGAAAACAUGGAUAAAGACAAAAGUCUGCUCUGUAUAAUGACCAGUGACAGAAUUUUGGAAUUUCAUUCCUGUUUCUUUCUGUAAAUAAUUUUCUUAUUUCUGAAAAGUACAUGGAAAUCAUGUAAAACCAUCAUUUCUGGGGUAAAGAGCAGUGACUUAUGUGCUCUUUUCCUUUUUUAGAUCCUGAGAG | ie_up |
| ENSG00000114316:-:3:49276426:49327816 | ENSG00000114316 | MSTRG.22947.1 | - | 3 | 49276227 | 49276436 | 210 | AAAAUUUAAGGUGAUUCUAAUUUUGAUGGUUUAACAGCUACAAUUAAGGGGAUUCUUGUUUCUGAAAAAGUUGUUGAAGAUCCUAGAUUUAGUUCAUGACAGUGCUCUUGCCUCCUCUUGGCAGCUCUGUGACUGCAUUUGGGGAGUUGACAGACAUUCUUCAGAAUCUGGCCUAGGAAACAGUUAUAUUUUGUGUAUAUGUUCCAGUUG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
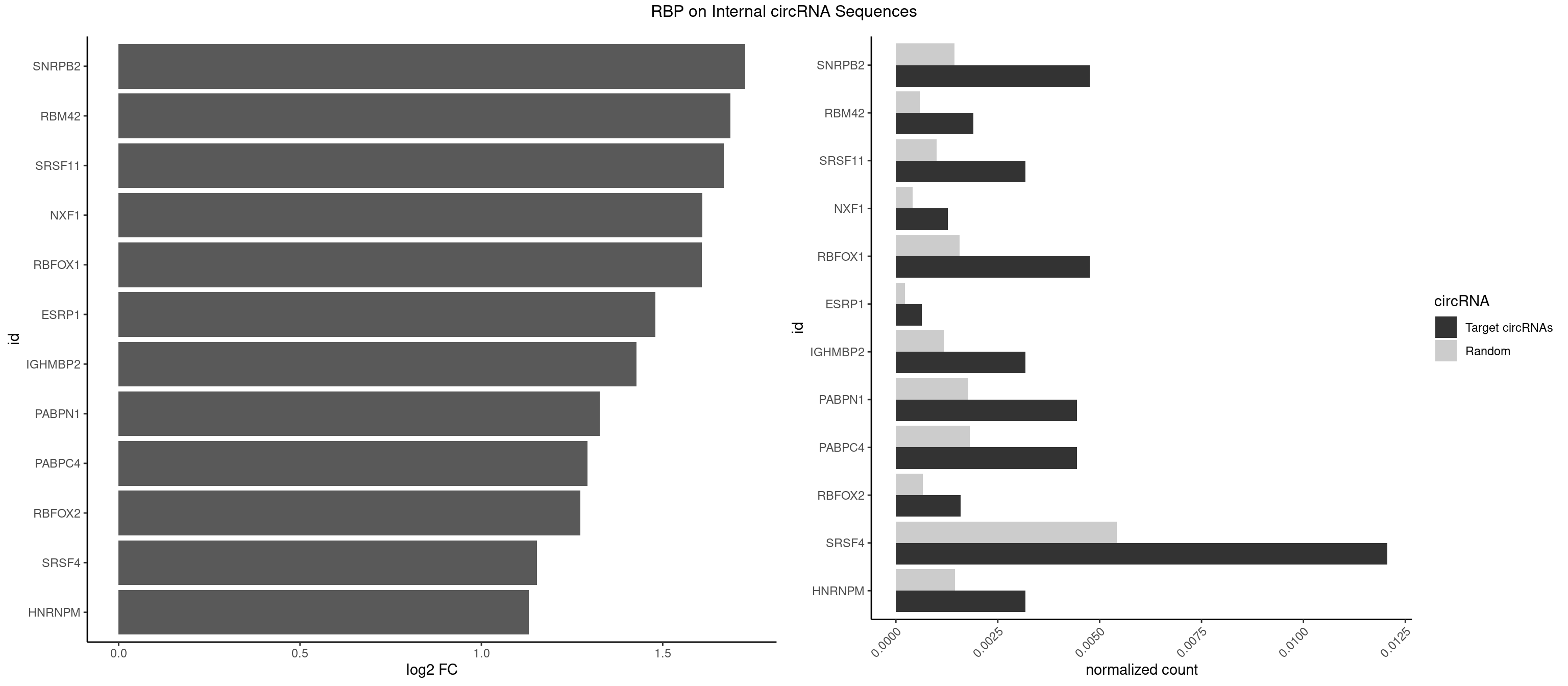
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRPB2 | 14 | 991 | 0.0047603935 | 0.0014376103 | 1.727408 | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM42 | 5 | 407 | 0.0019041574 | 0.0005912752 | 1.687251 | AACUAA,AACUAC,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| SRSF11 | 9 | 688 | 0.0031735957 | 0.0009985015 | 1.668282 | AAGAAG | AAGAAG |
| NXF1 | 3 | 286 | 0.0012694383 | 0.0004159215 | 1.609807 | AACCUG | AACCUG |
| RBFOX1 | 14 | 1077 | 0.0047603935 | 0.0015622419 | 1.607463 | AGCAUG,GCAUGA,GCAUGU,UGACUG,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| ESRP1 | 1 | 156 | 0.0006347191 | 0.0002275250 | 1.480093 | AGGGAU | AGGGAU |
| IGHMBP2 | 9 | 813 | 0.0031735957 | 0.0011796520 | 1.427757 | AAAAAA | AAAAAA |
| PABPN1 | 13 | 1222 | 0.0044430340 | 0.0017723764 | 1.325860 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| PABPC4 | 13 | 1251 | 0.0044430340 | 0.0018144033 | 1.292050 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBFOX2 | 4 | 452 | 0.0015867978 | 0.0006564894 | 1.273275 | UGACUG,UGCAUG | UGACUG,UGCAUG |
| SRSF4 | 37 | 3740 | 0.0120596636 | 0.0054214720 | 1.153433 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPM | 9 | 999 | 0.0031735957 | 0.0014492040 | 1.130858 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
RBP on BSJ (Exon Only)
Plot

Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383 | 1.531901 | AGAUCC | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
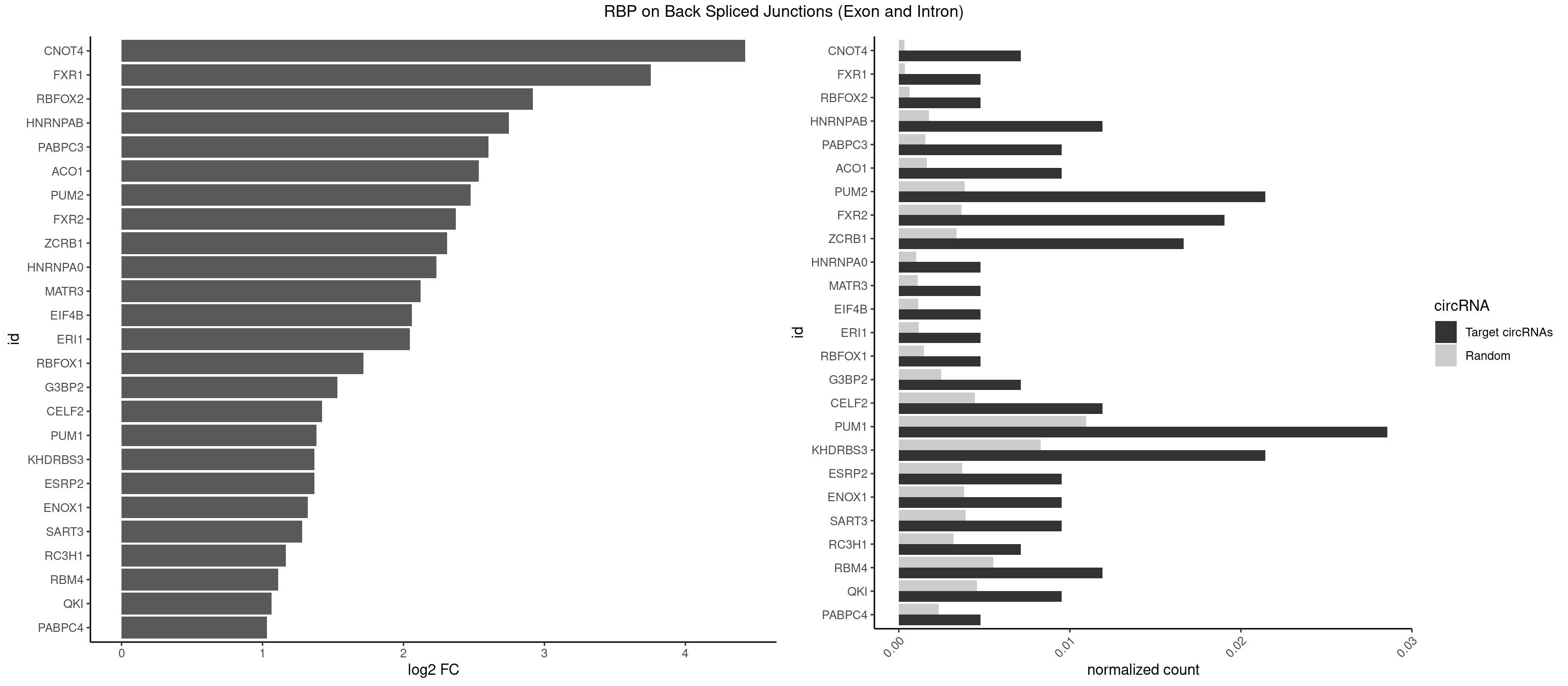
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 2 | 65 | 0.007142857 | 0.0003325272 | 4.424957 | GACAGA | GACAGA |
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| HNRNPAB | 4 | 351 | 0.011904762 | 0.0017734784 | 2.746885 | AAAGAC,AAGACA,AGACAA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC3 | 3 | 310 | 0.009523810 | 0.0015669085 | 2.603618 | AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ACO1 | 3 | 325 | 0.009523810 | 0.0016424829 | 2.535660 | CAGUGA,CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| PUM2 | 8 | 764 | 0.021428571 | 0.0038542926 | 2.474998 | GUAAAU,GUACAU,GUAUAU,UAAAUA,UGUAAA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| FXR2 | 7 | 730 | 0.019047619 | 0.0036829907 | 2.370661 | AGACAA,GACAAA,GACAGA,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| ZCRB1 | 6 | 666 | 0.016666667 | 0.0033605401 | 2.310201 | AAUUAA,AUUUAA,GACUUA,GAUUUA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUCAGA | UUCAGA,UUUCAG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | GGAUAA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| CELF2 | 4 | 881 | 0.011904762 | 0.0044437727 | 1.421682 | GUUGUU,UAUGUG,UAUGUU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PUM1 | 11 | 2172 | 0.028571429 | 0.0109482064 | 1.383879 | AGAAUU,CAGAAU,GUAAAU,GUACAU,GUAUAU,UAAAUA,UGUAAA,UGUAUA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGGAG,GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| KHDRBS3 | 8 | 1646 | 0.021428571 | 0.0082980653 | 1.368689 | AAAUAA,AUAAAG,GAUAAA,UAAAAC,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AAGACA,AGUACA,CAGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUCUU,CUCUUU,UUCCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | AAUCAU,ACUUAU,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAG | AAAAAA,AAAAAG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.