circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000184575:+:12:64409961:64430287
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000184575:+:12:64409961:64430287 | ENSG00000184575 | ENST00000332707 | + | 12 | 64409962 | 64430287 | 2050 | AUGUUUAUAAAUUCUUCUGUGGGAUCAGAGGGCACGCCUAUUACAACCAGAAAACUACAAGUAUAACAGCGAGGAUGGAUGAACAGGCUCUAUUAGGGCUAAAUCCAAAUGCUGAUUCAGACUUUAGACAAAGGGCCCUGGCCUAUUUUGAGCAGUUAAAAAUUUCCCCAGAUGCCUGGCAGGUGUGUGCAGAAGCUCUAGCCCAGAGGACAUACAGUGAUGAUCAUGUGAAGUUUUUCUGCUUUCAAGUACUGGAACAUCAAGUUAAAUACAAAUACUCAGAACUAACCACUGUUCAACAACAGCUAAUUAGGGAGACGCUCAUAUCAUGGCUGCAAGCUCAGAUGCUGAAUCCCCAACCAGAGAAGACCUUUAUACGAAAUAAAGCCGCCCAAGUCUUCGCCUUGCUUUUUGUUACAGAGUAUCUCACUAAGUGGCCCAAGUUUUUUUUUGACAUUCUCUCAGUAGUGGACCUAAAUCCAAGGGGAGUAGAUCUCUACCUGCGAAUCCUCAUGGCUAUUGAUUCAGAGUUGGUGGAUCGUGAUGUGGUGCAUACAUCAGAGGAGGCUCGUAGGAAUACUCUCAUAAAAGAUACCAUGAGGGAACAGUGCAUUCCAAAUCUGGUGGAAUCAUGGUACCAAAUAUUACAAAAUUAUCAGUUUACUAAUUCUGAAGUGACGUGUCAGUGCCUUGAAGUAGUUGGGGCUUAUGUCUCUUGGAUAGACUUAUCCCUUAUAGCCAAUGAUAGGUUUAUAAAUAUGCUGCUAGGUCAUAUGUCAAUAGAAGUUCUACGGGAAGAAGCAUGUGACUGUUUAUUUGAAGUUGUAAAUAAAGGAAUGGACCCUGUUGAUAAAAUGAAACUAGUGGAAUCUUUGUGUCAAGUAUUACAGUCUGCUGGGUUUUUCAGCAUUGACCAGGAAGAAGAUGUUGACUUCCUGGCCAGAUUUUCUAAGUUGGUAAAUGGAAUGGGACAGUCAUUGAUAGUUAGUUGGAGUAAAUUAAUUAAGAAUGGGGAUAUUAAGAAUGCUCAAGAGGCACUACAAGCUAUUGAAACAAAAGUGGCACUGAUGUUGCAGCUACUAAUUCAUGAGGAUGAUGAUAUUUCUUCUAAUAUUAUUGGAUUUUGUUACGAUUAUCUUCAUAUUUUGAAACAGCUUACAGUGCUCUCGGAUCAGCAAAAAGCUAAUGUAGAGGCAAUCAUGUUGGCCGUUAUGAAAAAAUUGACUUACGAUGAAGAAUAUAACUUUGAAAAUGAGGGUGAAGAUGAAGCCAUGUUUGUAGAAUAUAGAAAACAACUGAAGUUACUGUUGGACAGGCUUGCUCAAGUUUCACCAGAGUUACUACUGGCCUCUGUUCGCAGAGUUUUUAGUUCUACACUGCAGAAUUGGCAGACUACACGGUUUAUGGAAGUUGAAGUAGCAAUAAGAUUGCUGUAUAUGUUGGCAGAAGCUCUUCCAGUAUCUCAUGGUGCUCACUUCUCAGGUGAUGUUUCAAAAGCUAGUGCUUUGCAGGAUAUGAUGCGAACUCUGGUAACAUCAGGAGUCAGUUCCUAUCAGCAUACAUCUGUGACAUUGGAGUUCUUCGAAACUGUUGUUAGAUAUGAAAAGUUUUUCACAGUUGAACCUCAGCACAUUCCAUGUGUACUAAUGGCUUUCUUAGAUCACAGAGGUCUGCGGCAUUCCAGUGCAAAAGUUCGGAGCAGGACGGCUUACCUGUUUUCUAGAUUUGUCAAAUCUCUCAAUAAGCAAAUGAAUCCUUUCAUUGAGGAUAUUUUGAAUAGAAUACAAGAUUUAUUAGAGCUUUCUCCACCUGAGAAUGGCCACCAGUCCUUACUGAGCAGCGAUGAUCAACUUUUUAUUUAUGAGACAGCUGGAGUGCUGAUUGUUAAUAGUGAAUAUCCGGCAGAAAGGAAACAAGCCUUAAUGAGGAAUCUGUUGACUCCACUAAUGGAGAAGUUUAAAAUUCUGUUAGAAAAGUUGAUGCUGGCACAAGAUGAAGAAAGGCAAGCCUCUCUAGCAGACUGUCUUAACCAUGCUGUUGGAUUUGCAAGAUGUUUAUAAAUUCUUCUGUGGGAUCAGAGGGCACGCCUAUUACAACCAG | circ |
| ENSG00000184575:+:12:64409961:64430287 | ENSG00000184575 | ENST00000332707 | + | 12 | 64409962 | 64430287 | 22 | GGAUUUGCAAGAUGUUUAUAAA | bsj |
| ENSG00000184575:+:12:64409961:64430287 | ENSG00000184575 | ENST00000332707 | + | 12 | 64409762 | 64409971 | 210 | UACACAUUAGUGGAGUAGUUAGCAGAGCCCAUUUUUCUGAGAAUUAGGCUCUAACUUUUAGUUGUAAUUUAGAUAAGUUCCAAAAUAAGUUUUUAAACAUUUAAGCUUCUUUGCAUGAUUUUAUCCAUUUACGUUAUUCAGGAUAAGCAUCUAUUUGUUUAUCAAGUAAUGUUUUCUUUCAACUUUGUUUUUUGUGGCAGAUGUUUAUAA | ie_up |
| ENSG00000184575:+:12:64409961:64430287 | ENSG00000184575 | ENST00000332707 | + | 12 | 64430278 | 64430487 | 210 | GAUUUGCAAGGUAAGUGUGAUCACAGUUAAAAUUAUAAAGUGCAUGUAUCUACACAGACAGAACCACUUGUUUGGUGGGCACACACAUUUGUGUUUCCACAGUUGGAUAAUCCAGAGAAUUGUCUAAAUCUUUGUCCUCCAAAUCCAGUCAGUUAGAUUUGGUUGAAGAAGGGGAUAGGUUACAUACCAUUUACAUUUUUGCCUUGACAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
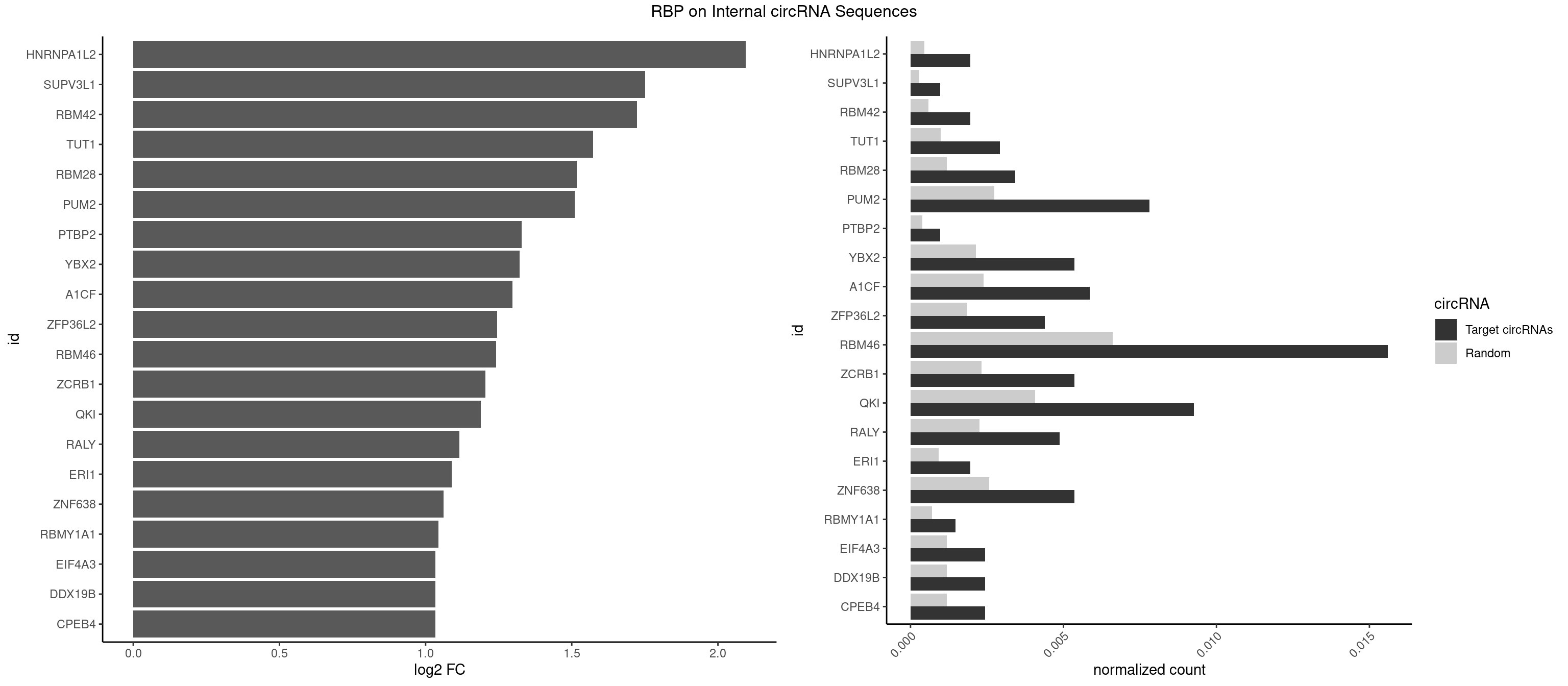
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 3 | 314 | 0.0019512195 | 0.0004564992 | 2.095692 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| SUPV3L1 | 1 | 199 | 0.0009756098 | 0.0002898408 | 1.751044 | CCGCCC | CCGCCC |
| RBM42 | 3 | 407 | 0.0019512195 | 0.0005912752 | 1.722474 | AACUAA,AACUAC,ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| TUT1 | 5 | 678 | 0.0029268293 | 0.0009840095 | 1.572594 | AAAUAC,AAUACU,AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM28 | 6 | 822 | 0.0034146341 | 0.0011926949 | 1.517506 | AGUAGA,AGUAGU,GAGUAG,UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PUM2 | 15 | 1890 | 0.0078048780 | 0.0027404447 | 1.509966 | GUAAAU,GUAGAU,GUAUAU,UAAAUA,UACAUC,UAGAUA,UGUAAA,UGUAGA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PTBP2 | 1 | 267 | 0.0009756098 | 0.0003883867 | 1.328811 | CUCUCU | CUCUCU |
| YBX2 | 10 | 1480 | 0.0053658537 | 0.0021462711 | 1.321975 | AACAAC,AACAUC,ACAACA,ACAACU,ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| A1CF | 11 | 1642 | 0.0058536585 | 0.0023810421 | 1.297745 | AGUAUA,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UCAGUA,UGAUCA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ZFP36L2 | 8 | 1277 | 0.0043902439 | 0.0018520827 | 1.245153 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBM46 | 31 | 4554 | 0.0156097561 | 0.0066011240 | 1.241664 | AAUCAU,AAUGAA,AAUGAU,AUCAAG,AUCAUG,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,GAUCAA,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZCRB1 | 10 | 1605 | 0.0053658537 | 0.0023274215 | 1.205075 | AAUUAA,GACUUA,GAUUUA,GGCUUA,GGUUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| QKI | 18 | 2805 | 0.0092682927 | 0.0040664663 | 1.188528 | AAUCAU,ACUAAC,ACUAAU,ACUUAU,CACUAA,CUAACC,CUCAUA,UACUCA,UCAUAU,UCUAAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RALY | 9 | 1553 | 0.0048780488 | 0.0022520629 | 1.115057 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ERI1 | 3 | 632 | 0.0019512195 | 0.0009173461 | 1.088838 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| ZNF638 | 10 | 1773 | 0.0053658537 | 0.0025708878 | 1.061541 | GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBMY1A1 | 2 | 489 | 0.0014634146 | 0.0007101099 | 1.043224 | ACAAGA | ACAAGA,CAAGAC |
| CPEB4 | 4 | 821 | 0.0024390244 | 0.0011912456 | 1.033833 | UUUUUU | UUUUUU |
| DDX19B | 4 | 821 | 0.0024390244 | 0.0011912456 | 1.033833 | UUUUUU | UUUUUU |
| EIF4A3 | 4 | 821 | 0.0024390244 | 0.0011912456 | 1.033833 | UUUUUU | UUUUUU |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
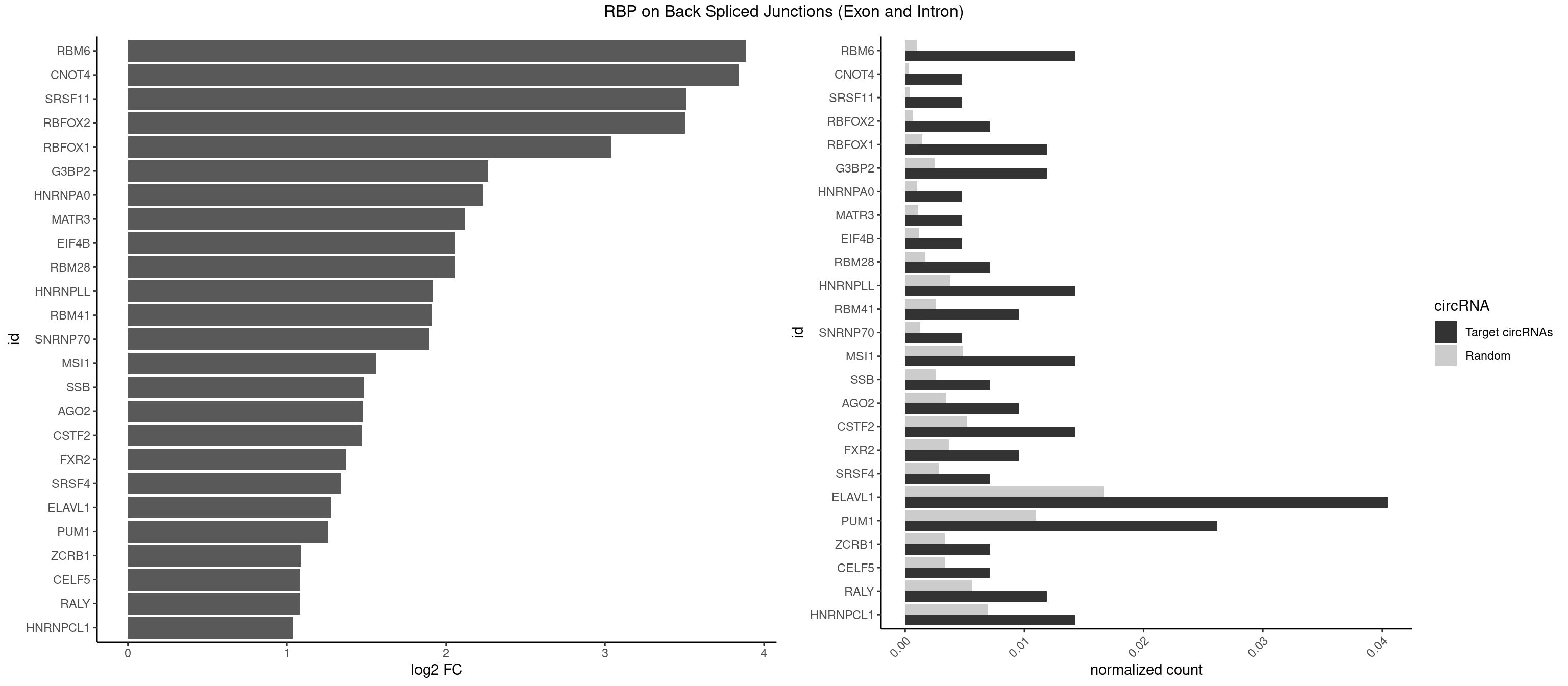
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM6 | 5 | 191 | 0.014285714 | 0.0009673519 | 3.884389 | AAUCCA,AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| RBFOX2 | 2 | 124 | 0.007142857 | 0.0006297864 | 3.503567 | UGCAUG | UGACUG,UGCAUG |
| RBFOX1 | 4 | 287 | 0.011904762 | 0.0014510278 | 3.036392 | GCAUGA,GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| G3BP2 | 4 | 490 | 0.011904762 | 0.0024738009 | 2.266737 | AGGAUA,GGAUAA,GGAUAG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGU,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPLL | 5 | 748 | 0.014285714 | 0.0037736800 | 1.920529 | ACACAC,ACAUAC,CACACA,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | UACAUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| MSI1 | 5 | 961 | 0.014285714 | 0.0048468360 | 1.559458 | AGUUAG,AGUUGG,UAGUUA,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| AGO2 | 3 | 677 | 0.009523810 | 0.0034159613 | 1.479247 | AAAGUG,AAGUGC,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUUU,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| FXR2 | 3 | 730 | 0.009523810 | 0.0036829907 | 1.370661 | AGACAG,GACAGA,UGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AAGAAG,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ELAVL1 | 16 | 3309 | 0.040476190 | 0.0166767432 | 1.279236 | UGAUUU,UGUUUU,UUAGUU,UUGGUU,UUGUUU,UUUAGU,UUUGGU,UUUGUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| PUM1 | 10 | 2172 | 0.026190476 | 0.0109482064 | 1.258348 | AAUGUU,AAUUGU,AGAAUU,AGAUAA,GAAUUG,UAAUGU,UACAUA,UAGAUA,UGUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AUUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 | 5 | 1381 | 0.014285714 | 0.0069629182 | 1.036809 | AUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.