circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000166225:+:12:69530864:69532056
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000166225:+:12:69530864:69532056 | ENSG00000166225 | MSTRG.7651.2 | + | 12 | 69530865 | 69532056 | 139 | AAAUGACCAUUAAGAAGUAGAUGCCCAGAUGCAAAAGUGAUGAAACAGUCCAUUUGUCAUAAAGUAAGAUGCAGCUGUGGCAUGUCAACCAGCUUGGAACAAAAUUGUAUCUGUUUUUCUCAGAAGAGAAUUCCACAAGAAAUGACCAUUAAGAAGUAGAUGCCCAGAUGCAAAAGUGAUGAAACAGUC | circ |
| ENSG00000166225:+:12:69530864:69532056 | ENSG00000166225 | MSTRG.7651.2 | + | 12 | 69530865 | 69532056 | 22 | AAUUCCACAAGAAAUGACCAUU | bsj |
| ENSG00000166225:+:12:69530864:69532056 | ENSG00000166225 | MSTRG.7651.2 | + | 12 | 69530665 | 69530874 | 210 | CCCAUGAGAUUGAGGCUGCAGUGAGGCGUGAUGGUGCCACUGCGCUUCAGCCUGAGCGACAGAAUGAGACCCUGUCUCAAAGGGGAAAAAAGUGCUUUGAGAGUUCUUUCUGGUCACUACAAUUUAGAUAAUUAUUCUUAAAGAGUUUUGUUAUAAUAUAUUCAAGAGUAACAUUCUUAAUAACUUAACCUAUAUUUCAGAAAUGACCAU | ie_up |
| ENSG00000166225:+:12:69530864:69532056 | ENSG00000166225 | MSTRG.7651.2 | + | 12 | 69532047 | 69532256 | 210 | AUUCCACAAGGUAAGAAAUUGUAAAUAUAGUAAUAAAACUAAUAUUGUUGAGUAAGGCUUAAUUGCUCCUGAUUUUUAAAAAUUUGUUUCCAUGUAUUUAUUUAAUAUCUUUUCUUUAUUAUGGGAUUCUCACUAAUUUAUUAGCCACUGUUUUACUAUGUCAGAAUUUGCUAGCAAGACACAGAUCAUCUUGUUUGAAUUUAGGAUAAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
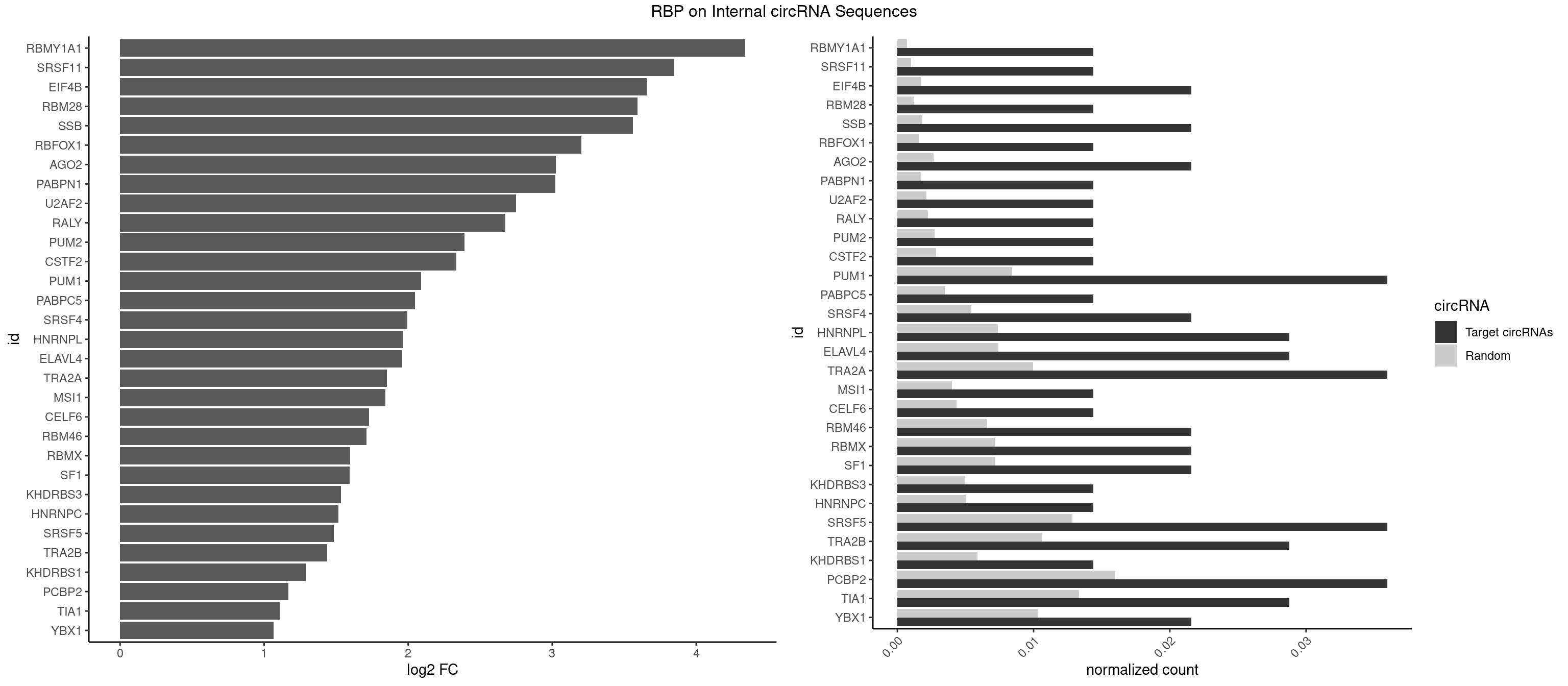
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 489 | 0.01438849 | 0.0007101099 | 4.340729 | ACAAGA | ACAAGA,CAAGAC |
| SRSF11 | 1 | 688 | 0.01438849 | 0.0009985015 | 3.849007 | AAGAAG | AAGAAG |
| EIF4B | 2 | 1179 | 0.02158273 | 0.0017100607 | 3.657758 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 | 1 | 822 | 0.01438849 | 0.0011926949 | 3.592618 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SSB | 2 | 1260 | 0.02158273 | 0.0018274462 | 3.561977 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBFOX1 | 1 | 1077 | 0.01438849 | 0.0015622419 | 3.203225 | GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| AGO2 | 2 | 1830 | 0.02158273 | 0.0026534924 | 3.023913 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPN1 | 1 | 1222 | 0.01438849 | 0.0017723764 | 3.021158 | AGAAGA | AAAAGA,AGAAGA |
| U2AF2 | 1 | 1477 | 0.01438849 | 0.0021419234 | 2.747936 | UUUUUC | UUUUCC,UUUUUC,UUUUUU |
| RALY | 1 | 1553 | 0.01438849 | 0.0022520629 | 2.675596 | UUUUUC | UUUUUC,UUUUUG,UUUUUU |
| PUM2 | 1 | 1890 | 0.01438849 | 0.0027404447 | 2.392433 | GUAGAU | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| CSTF2 | 1 | 1967 | 0.01438849 | 0.0028520334 | 2.334852 | UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| PUM1 | 4 | 5823 | 0.03597122 | 0.0084401638 | 2.091500 | AAUUGU,AGAAUU,AUUGUA,GUAGAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| PABPC5 | 1 | 2400 | 0.01438849 | 0.0034795387 | 2.047947 | AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SRSF4 | 2 | 3740 | 0.02158273 | 0.0054214720 | 1.993121 | AAGAAG,AGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPL | 3 | 5085 | 0.02877698 | 0.0073706513 | 1.965051 | AACAAA,CACAAG,CAUAAA | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| ELAVL4 | 3 | 5105 | 0.02877698 | 0.0073996354 | 1.959389 | GUAUCU,UGUAUC,UUGUAU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TRA2A | 4 | 6871 | 0.03597122 | 0.0099589296 | 1.852781 | AAGAAA,AAGAAG,AGAAGA,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| MSI1 | 1 | 2770 | 0.01438849 | 0.0040157442 | 1.841176 | AGUAAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| CELF6 | 1 | 2997 | 0.01438849 | 0.0043447134 | 1.727582 | GUGAUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM46 | 2 | 4554 | 0.02158273 | 0.0066011240 | 1.709094 | AUGAAA,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBMX | 2 | 4925 | 0.02158273 | 0.0071387787 | 1.596128 | AAGAAG,AAGUAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| SF1 | 2 | 4931 | 0.02158273 | 0.0071474739 | 1.594372 | ACAGUC,AGUAAG | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| KHDRBS3 | 1 | 3429 | 0.01438849 | 0.0049707696 | 1.533374 | AUAAAG | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPC | 1 | 3471 | 0.01438849 | 0.0050316361 | 1.515816 | UUUUUC | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SRSF5 | 4 | 8869 | 0.03597122 | 0.0128544391 | 1.484577 | AAGAAG,AGAAGA,AUAAAG,UGCAGC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| TRA2B | 3 | 7329 | 0.02877698 | 0.0106226650 | 1.437769 | AAGAAG,AGAAGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| KHDRBS1 | 1 | 4064 | 0.01438849 | 0.0058910141 | 1.288327 | CAAAAU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| PCBP2 | 4 | 11045 | 0.03597122 | 0.0160079069 | 1.168059 | AAUUCC,AUUCCA,CAUUAA,CCAUUA | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| TIA1 | 3 | 9206 | 0.02877698 | 0.0133428208 | 1.108851 | GUUUUU,UUUUCU,UUUUUC | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| YBX1 | 2 | 7119 | 0.02158273 | 0.0103183321 | 1.064668 | CAACCA,CCACAA | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
RBP on BSJ (Exon Only)
Plot
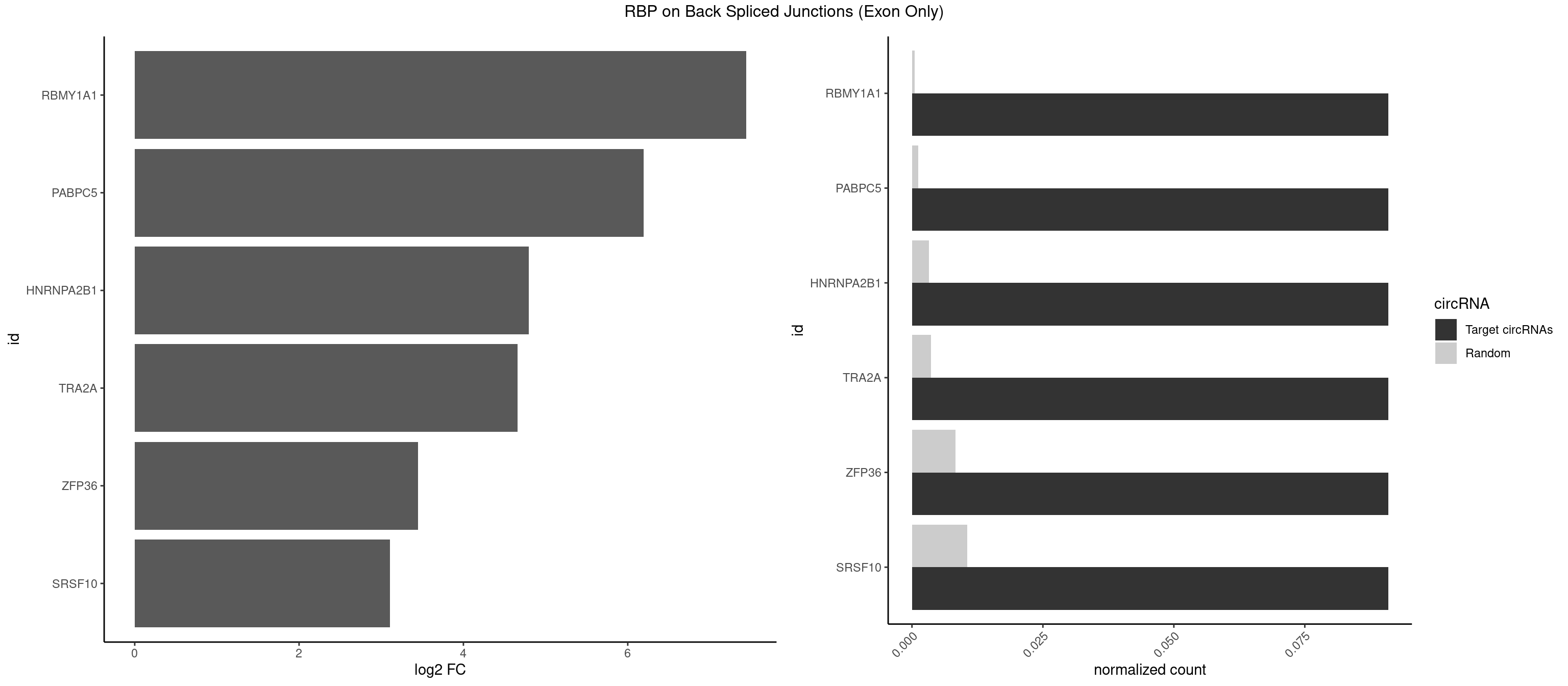
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBMY1A1 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | ACAAGA | ACAAGA,CAAGAC |
| PABPC5 | 1 | 18 | 0.09090909 | 0.0012444328 | 6.190864 | AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.0032748232 | 4.794936 | CAAGAA | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| TRA2A | 1 | 54 | 0.09090909 | 0.0036023055 | 4.657432 | AAGAAA | AAAGAA,AAGAAA,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | AAGAAA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
| SRSF10 | 1 | 160 | 0.09090909 | 0.0105449306 | 3.107875 | AAGAAA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
RBP on BSJ (Exon and Intron)
Plot
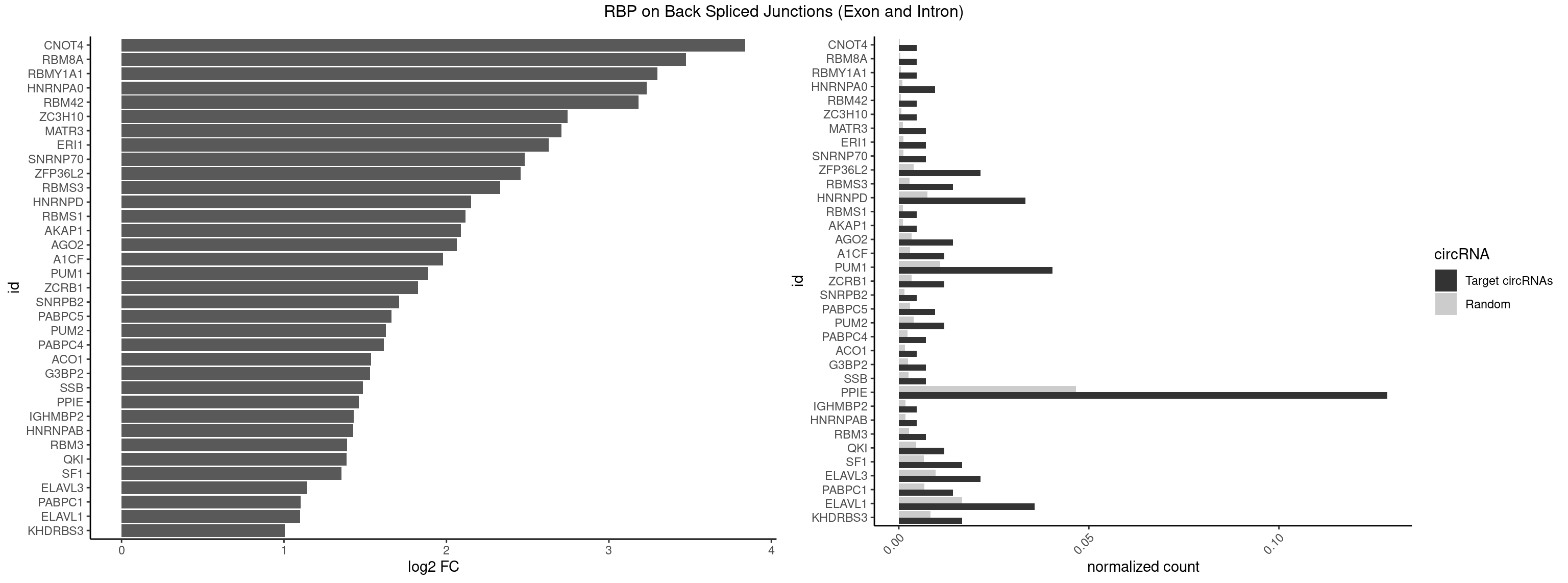
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CNOT4 | 1 | 65 | 0.004761905 | 0.0003325272 | 3.839994 | GACAGA | GACAGA |
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | UGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| HNRNPA0 | 3 | 200 | 0.009523810 | 0.0010126965 | 3.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | GAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ZFP36L2 | 8 | 774 | 0.021428571 | 0.0039046755 | 2.456261 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RBMS3 | 5 | 562 | 0.014285714 | 0.0028365578 | 2.332360 | AAUAUA,AUAUAG,AUAUAU,CUAUAU | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPD | 13 | 1488 | 0.033333333 | 0.0075020153 | 2.151615 | AAAAAA,AAUUUA,AUUUAU,UAUUUA,UUAGGA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| AGO2 | 5 | 677 | 0.014285714 | 0.0034159613 | 2.064210 | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,UAAUUA,UAAUUG,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| PUM1 | 16 | 2172 | 0.040476190 | 0.0109482064 | 1.886379 | AAUAUU,AAUUGU,AGAAUU,AGAUAA,AUUGUA,CAGAAU,GUAAAU,GUAAUA,UAAAUA,UAAUAU,UAGAUA,UGUAAA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ZCRB1 | 4 | 666 | 0.011904762 | 0.0033605401 | 1.824774 | ACUUAA,AUUUAA,GCUUAA,GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUAAAU,UAAAUA,UAGAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| G3BP2 | 2 | 490 | 0.007142857 | 0.0024738009 | 1.529772 | AGGAUA,GGAUAA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| PPIE | 53 | 9262 | 0.128571429 | 0.0466696896 | 1.462012 | AAAAAA,AAAAAU,AAAAUU,AAAUAU,AAAUUU,AAUAAA,AAUAUA,AAUAUU,AAUUAU,AAUUUA,AUAAAA,AUAAUA,AUAAUU,AUAUAU,AUAUUU,AUUAUU,AUUUAA,AUUUAU,AUUUUU,UAAAAA,UAAAUA,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAU,UAUAUU,UAUUAU,UAUUUA,UUAAAA,UUAAUA,UUAAUU,UUAUAA,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUU,UUUUAA,UUUUUA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AAGACA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,AAACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACUAAU,CACUAA,UAACCU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | ACUAAU,AGUAAC,AGUAAG,CACAGA,UAGUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| ELAVL3 | 8 | 1928 | 0.021428571 | 0.0097188634 | 1.140676 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| PABPC1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | AAAAAA,ACUAAU,CUAAUA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| ELAVL1 | 14 | 3309 | 0.035714286 | 0.0166767432 | 1.098664 | AUUUAU,UAUUUA,UGAUUU,UGUUUU,UUAUUU,UUGUUU,UUUAUU,UUUGUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 0.0082980653 | 1.006119 | AAUAAA,AGAUAA,AUAAAA,GAUAAA,UAAAAC,UAAAUA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.