circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000141556:+:17:82797756:82884202
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000141556:+:17:82797756:82884202 | ENSG00000141556 | ENST00000681983 | + | 17 | 82797757 | 82884202 | 762 | GCACAAAUAUUUAAACAUGGAAAACGUGAAGACUGUUUGCCCUAUGCUGCCACUGUCCUCAGGUGCCUCGAUGGCUGCAGACUCCCUGAGAGCAACCAGACCCUGCUGCGGAAGCUGGGGGUGAAGCUUGUGCAGCGACUGGGGCUGACAUUCCUGAAGCCGAAGGUGGCAGCAUGGAGGUACCAGCGUGGCUGCCGAUCUUUGGCUGCAAAUCUGCAGCUCCUCACUCAGGGUCAGAGUGAGCAGAAGCCACUCAUCCUGACCGAAGAUGACGACGAAGAUGACGACGUCCCAGAGGGGGUGGAGCGUGUGAUAGAGCAGCUGCUGGUCGGGCUGAAGGACAAGGACACGGUCGUGCGGUGGUCUGCAGCCAAGGGCAUCGGUAGGAUGGCUGGCAGGCUUCCCAGAGCCCUGGCGGAUGAUGUGGUCGGGUCUGUGCUGGACUGCUUCAGUUUCCAGGAGACUGACAAGGCGUGGCAUGGGGGAUGUCUGGCGCUGGCAGAGCUGGGCAGGAGAGGCCUGUUGCUGCCGUCUCGACUCGUGGAUGUUGUCGCCGUGAUCCUGAAGGCGCUGACCUACGACGAGAAGCGGGGUGCCUGCAGCGUGGGCACCAACGUCAGGGACGCCGCCUGCUACGUGUGCUGGGCCUUCGCGCGUGCCUAUGAGCCUCAGGAGCUGAAGCCCUUUGUGACUGCAAUCUCGAGUGCACUGGUGAUUGCUGCGGUGUUUGACCGAGACAUAAACUGCAGAAGAGCAGCCUCUGCACAAAUAUUUAAACAUGGAAAACGUGAAGACUGUUUGCCCUAUGCUGC | circ |
| ENSG00000141556:+:17:82797756:82884202 | ENSG00000141556 | ENST00000681983 | + | 17 | 82797757 | 82884202 | 22 | GAGCAGCCUCUGCACAAAUAUU | bsj |
| ENSG00000141556:+:17:82797756:82884202 | ENSG00000141556 | ENST00000681983 | + | 17 | 82797557 | 82797766 | 210 | AGAUGAAUUUUUGGGAGAUGAUUUCACUAGUCUCUUAUAAUUACUUUAUCAUGUACAGAAACUGUUUUCUUUAUUAUUAGGUGGUGAGUGUUUAGAAUAAAAAUUCUGAAAGAUGUGAUGAAUUGUUUUCACAAUUUGUGUGUAUGUUUUAAACUUCUAUUUGUAUUUGUAACAUUAAAAAUCUUUUUUUUUUUUUUUAGGCACAAAUAU | ie_up |
| ENSG00000141556:+:17:82797756:82884202 | ENSG00000141556 | ENST00000681983 | + | 17 | 82884193 | 82884402 | 210 | AGCAGCCUCUGUAAGUUUUCUCAUUUUGAUAUUUCCUUUCCUGAAGGUGGGGGGUGGGCCUGGUCUCCCUGAUGCUCCUCUGUGCACUGCUGCCUGGCCAGCUCACCCAUCCACAGCAGGAGCCGCGAGGGAGCUGCUGAGAGUGCCAGCUGCGGGCAGGCCACUGGUUCUUACUGUCUCUGGGCGUCUUCAGCGUGUGAAGGGCGGUCA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
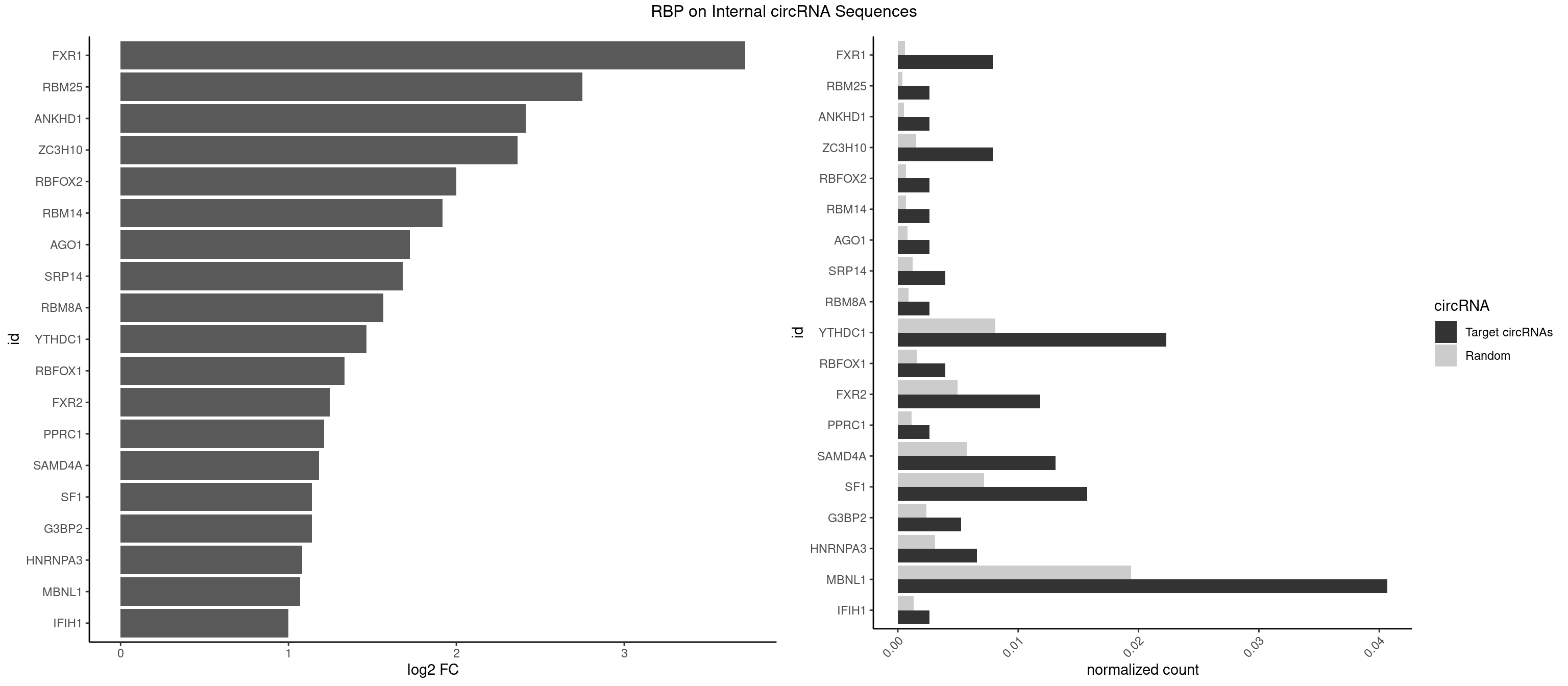
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 5 | 411 | 0.007874016 | 0.0005970720 | 3.721123 | ACGACG,AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM25 | 1 | 268 | 0.002624672 | 0.0003898359 | 2.751198 | UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| ANKHD1 | 1 | 339 | 0.002624672 | 0.0004927293 | 2.413270 | GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| ZC3H10 | 5 | 1053 | 0.007874016 | 0.0015274610 | 2.365964 | CAGCGA,CCAGCG,GCAGCG,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBFOX2 | 1 | 452 | 0.002624672 | 0.0006564894 | 1.999293 | UGACUG | UGACUG,UGCAUG |
| RBM14 | 1 | 478 | 0.002624672 | 0.0006941687 | 1.918779 | CGCGCG | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| AGO1 | 1 | 548 | 0.002624672 | 0.0007956130 | 1.721998 | GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SRP14 | 2 | 847 | 0.003937008 | 0.0012289250 | 1.679703 | CGCCUG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM8A | 1 | 611 | 0.002624672 | 0.0008869128 | 1.565273 | CGCGCG | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| YTHDC1 | 16 | 5576 | 0.022309711 | 0.0080822104 | 1.464850 | GACUGC,GAGUGC,GCCUGC,GCGUGC,GGCUGC,GGGUGC,UCGUGC,UGCUAC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| RBFOX1 | 2 | 1077 | 0.003937008 | 0.0015622419 | 1.333482 | AGCAUG,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| FXR2 | 8 | 3434 | 0.011811024 | 0.0049780156 | 1.246491 | GACAAG,GACGAA,GACGAG,GGACAA,UGACAA,UGACGA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| PPRC1 | 1 | 780 | 0.002624672 | 0.0011318283 | 1.213482 | CGCGCG | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| SAMD4A | 9 | 3992 | 0.013123360 | 0.0057866714 | 1.181331 | CGGGUC,CUGGAC,CUGGCA,CUGGUC,GCUGGA,GCUGGC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SF1 | 11 | 4931 | 0.015748031 | 0.0071474739 | 1.139666 | ACUGAC,CGCUGA,GCUGAC,GCUGCC,UGCUGC | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| G3BP2 | 3 | 1644 | 0.005249344 | 0.0023839405 | 1.138789 | AGGAUG,GGAUGA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPA3 | 4 | 2140 | 0.006561680 | 0.0031027457 | 1.080520 | AGGAGC,CAAGGA,CCAAGG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| MBNL1 | 30 | 13372 | 0.040682415 | 0.0193802045 | 1.069822 | CCUGCU,CUGCCA,CUGCCG,CUGCGG,CUGCUA,CUGCUG,CUGCUU,CUUGUG,GCGCUG,GCUGCG,GCUGCU,GCUUGU,GUCUCG,GUGCUG,UGCUGC,UGCUUC,UUGCCC,UUGCUG,UUGUGC | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| IFIH1 | 1 | 904 | 0.002624672 | 0.0013115296 | 1.000887 | GCGGAU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
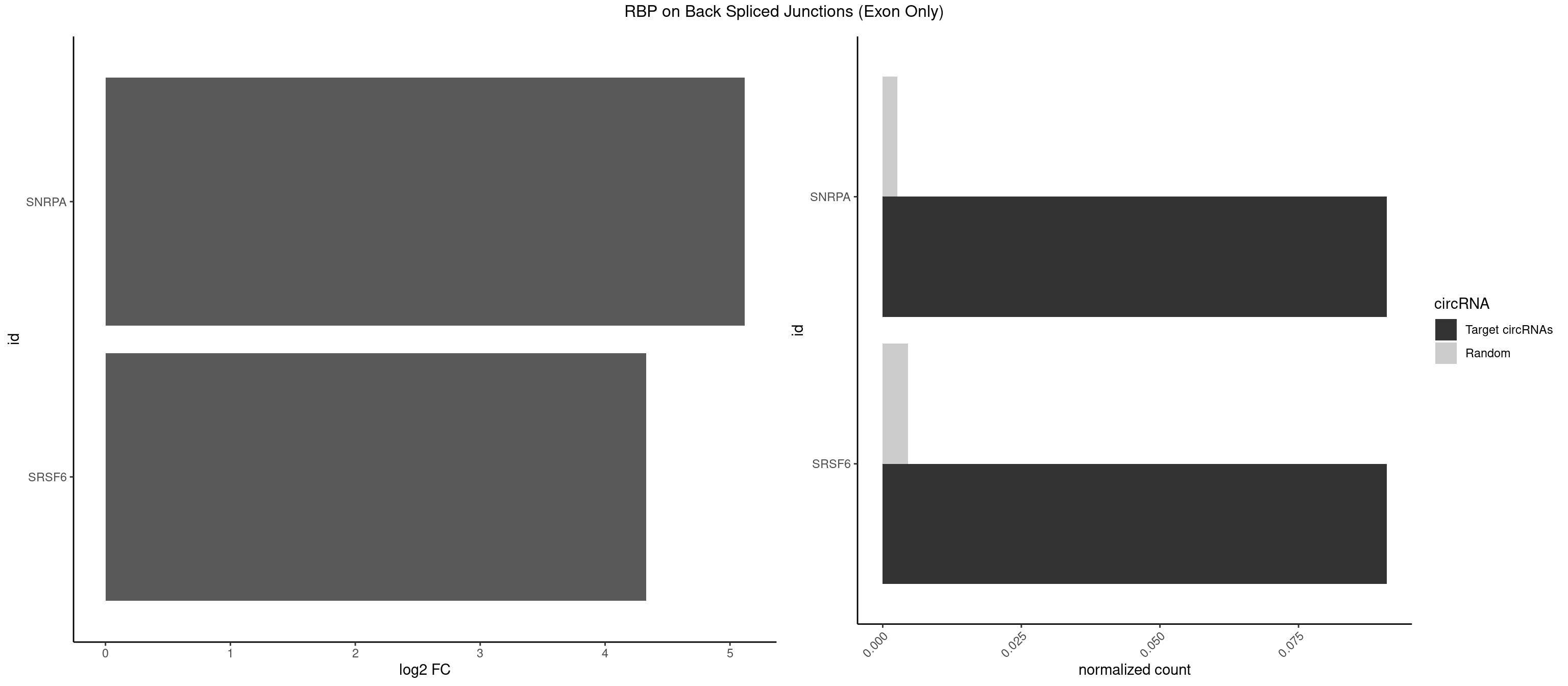
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRPA | 1 | 39 | 0.09090909 | 0.002619859 | 5.116864 | UGCACA | AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,GAGCAG,GAUACC,GCAGUA,GGAGAU,GGGUAU,GUAGGC,GUAGUG,UAUGCU,UCCUGC,UGCACG,UUCCUG,UUGCAC |
| SRSF6 | 1 | 68 | 0.09090909 | 0.004519256 | 4.330267 | CCUCUG | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
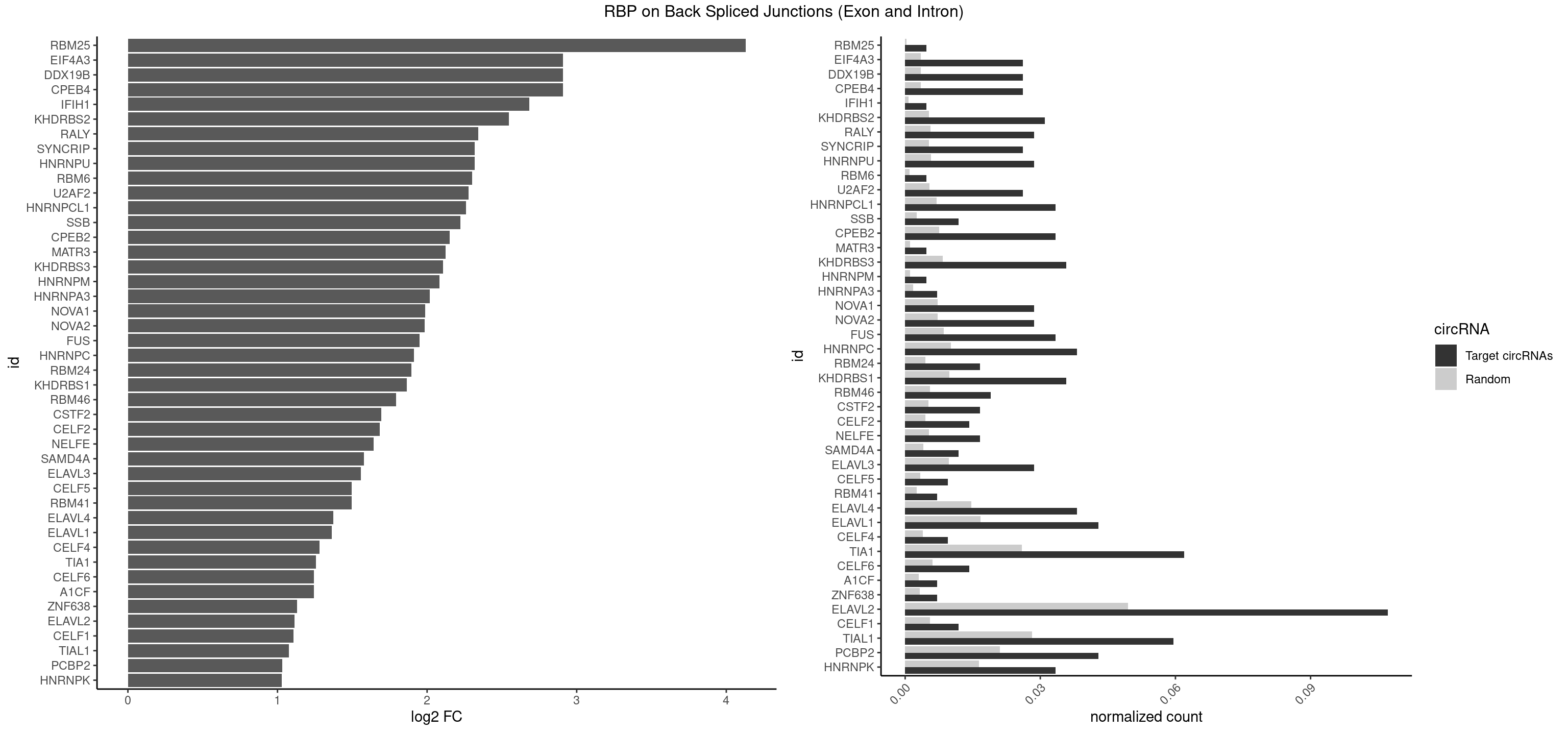
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| CPEB4 | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| DDX19B | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| EIF4A3 | 10 | 691 | 0.026190476 | 0.0034864974 | 2.909192 | UUUUUU | UUUUUU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GCCGCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| KHDRBS2 | 12 | 1051 | 0.030952381 | 0.0053002821 | 2.545909 | AAUAAA,AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RALY | 11 | 1117 | 0.028571429 | 0.0056328094 | 2.342647 | UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| SYNCRIP | 10 | 1041 | 0.026190476 | 0.0052498992 | 2.318681 | UUUUUU | AAAAAA,UUUUUU |
| HNRNPU | 11 | 1136 | 0.028571429 | 0.0057285369 | 2.318335 | GGGGGG,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| RBM6 | 1 | 191 | 0.004761905 | 0.0009673519 | 2.299426 | CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| U2AF2 | 10 | 1071 | 0.026190476 | 0.0054010480 | 2.277731 | UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| HNRNPCL1 | 13 | 1381 | 0.033333333 | 0.0069629182 | 2.259202 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| SSB | 4 | 505 | 0.011904762 | 0.0025493753 | 2.223323 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| CPEB2 | 13 | 1487 | 0.033333333 | 0.0074969770 | 2.152585 | AUUUUU,CAUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| KHDRBS3 | 14 | 1646 | 0.035714286 | 0.0082980653 | 2.105654 | AAUAAA,AUAAAA,AUUAAA,UUAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AGGAGC,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| NOVA1 | 11 | 1428 | 0.028571429 | 0.0071997179 | 1.988561 | GGGGGG,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| NOVA2 | 11 | 1434 | 0.028571429 | 0.0072299476 | 1.982516 | GGGGGG,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| FUS | 13 | 1711 | 0.033333333 | 0.0086255542 | 1.950277 | GGGGGG,GGGUGG,UGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPC | 15 | 2006 | 0.038095238 | 0.0101118501 | 1.913564 | AUUUUU,CUUUUU,GGGGGG,UUUUUA,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RBM24 | 6 | 888 | 0.016666667 | 0.0044790407 | 1.895704 | AGAGUG,GAGUGU,GUGUGA,GUGUGU,UGAGUG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| KHDRBS1 | 14 | 1945 | 0.035714286 | 0.0098045143 | 1.864983 | AUAAAA,UAAAAA,UUAAAA,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBM46 | 7 | 1091 | 0.019047619 | 0.0055018138 | 1.791631 | AUCAUG,AUGAAU,AUGAUU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CSTF2 | 6 | 1022 | 0.016666667 | 0.0051541717 | 1.693153 | GUGUGU,GUGUUU,UGUGUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF2 | 5 | 881 | 0.014285714 | 0.0044437727 | 1.684716 | GUAUGU,GUGUGU,UAUGUU,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| NELFE | 6 | 1059 | 0.016666667 | 0.0053405885 | 1.641895 | CUCUGG,CUGGUU,GGUCUC,GUCUCU,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SAMD4A | 4 | 790 | 0.011904762 | 0.0039852882 | 1.578783 | CGGGCA,CUGGCC,CUGGUC,GCGGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| ELAVL3 | 11 | 1928 | 0.028571429 | 0.0097188634 | 1.555714 | UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGU,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ELAVL4 | 15 | 2916 | 0.038095238 | 0.0146966949 | 1.374119 | UUGUAU,UUUAUU,UUUGUA,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| ELAVL1 | 17 | 3309 | 0.042857143 | 0.0166767432 | 1.361698 | UGAUUU,UGUUUU,UUGUUU,UUUAUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GUGUGU,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| TIA1 | 25 | 5140 | 0.061904762 | 0.0259018541 | 1.256995 | AUUUUG,AUUUUU,CCUUUC,CUUUUU,GUUUUA,GUUUUC,UCCUUU,UUUAUU,UUUUCU,UUUUGG,UUUUUA,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CELF6 | 5 | 1196 | 0.014285714 | 0.0060308343 | 1.244144 | GUGAUG,GUGGGG,GUGGUG,UGUGAU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUAAUU,UAAUUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| ELAVL2 | 44 | 9826 | 0.107142857 | 0.0495112858 | 1.113706 | AAUUUG,AAUUUU,AUAUUU,AUUAUU,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUUU,GUUUUA,GUUUUC,UAAGUU,UACUUU,UAUGUU,UAUUAU,UAUUUG,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUACUU,UUCUUU,UUGUAU,UUUAUU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUUA,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | CUGUCU,GUGUGU,UGUGUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| TIAL1 | 24 | 5597 | 0.059523810 | 0.0282043531 | 1.077549 | AAUUUU,AGUUUU,AUUUUG,AUUUUU,CUUUUU,GUUUUC,UUUAAA,UUUAUU,UUUUAA,UUUUCA,UUUUUA,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PCBP2 | 17 | 4164 | 0.042857143 | 0.0209844821 | 1.030213 | AAAUUC,AAUUAC,ACAUUA,AUUAAA,CAUUAA,CUCCCU,GGGGGG,UUUUUU | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| HNRNPK | 13 | 3244 | 0.033333333 | 0.0163492543 | 1.027741 | ACCCAU,CCAUCC,GGGGGG,UUUUUU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.