circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000159692:-:4:1241324:1241519
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000159692:-:4:1241324:1241519 | ENSG00000159692 | ENST00000382952 | - | 4 | 1241325 | 1241519 | 195 | UAAGACUUUGACCCCGACGCAGGCUUCAGUGAUUGCCGCGGAUCAGAAGCCUGAGCACUUUGGCUCGAGACAAUUAAGGACGUGGGAUGAGGCUCCGAGACAGGACGCGGUUCUGCCUGGGGAUCCUGAAGAUAAAAAGCUUUGAAAAGUCGAAUUCAUGGUCGUGGAAGCUGAGCCCAUAUUAAGAGAUGUCAGUAAGACUUUGACCCCGACGCAGGCUUCAGUGAUUGCCGCGGAUCAGAAGC | circ |
| ENSG00000159692:-:4:1241324:1241519 | ENSG00000159692 | ENST00000382952 | - | 4 | 1241325 | 1241519 | 22 | AGAGAUGUCAGUAAGACUUUGA | bsj |
| ENSG00000159692:-:4:1241324:1241519 | ENSG00000159692 | ENST00000382952 | - | 4 | 1241510 | 1241719 | 210 | AGGGUGAGCGCCCUUCGUGUGUGCGGCAGGAGCCGGGGCCCCUGGCUGUCAGCAGGCCCCCGAGUCCAGGUAGGUCCCAGACAGCAGUCCCGGGUGUGGCAGAUGCAGUGAGGUCCACUUCUGAGGCGUUCCCUGGAGUCUGUGAGUUCUGGUCGGACCUUCGAUGGCAUUUUAAUGUGUCUCUUGUGUUUGAUUUUUAGUAAGACUUUG | ie_up |
| ENSG00000159692:-:4:1241324:1241519 | ENSG00000159692 | ENST00000382952 | - | 4 | 1241125 | 1241334 | 210 | GAGAUGUCAGGUUGGUAGGGGUUCCUGGCGUCGGCGGCAGAGUGAAGCCGGCCGGUCUCUUUGCGACCGUGUGGAGGAGGCACUGCCUCGGAUCAAGUCGAGGGACCUGGGCUGCAUGGGCGUCGGGCCGGACUGCUGCCACCUGACCCCCGGUGUGCUCUGGGCGAGGUCAGGUGCCCCUCGAGGGAGGGCUUCCUGCACACCUGGUGG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
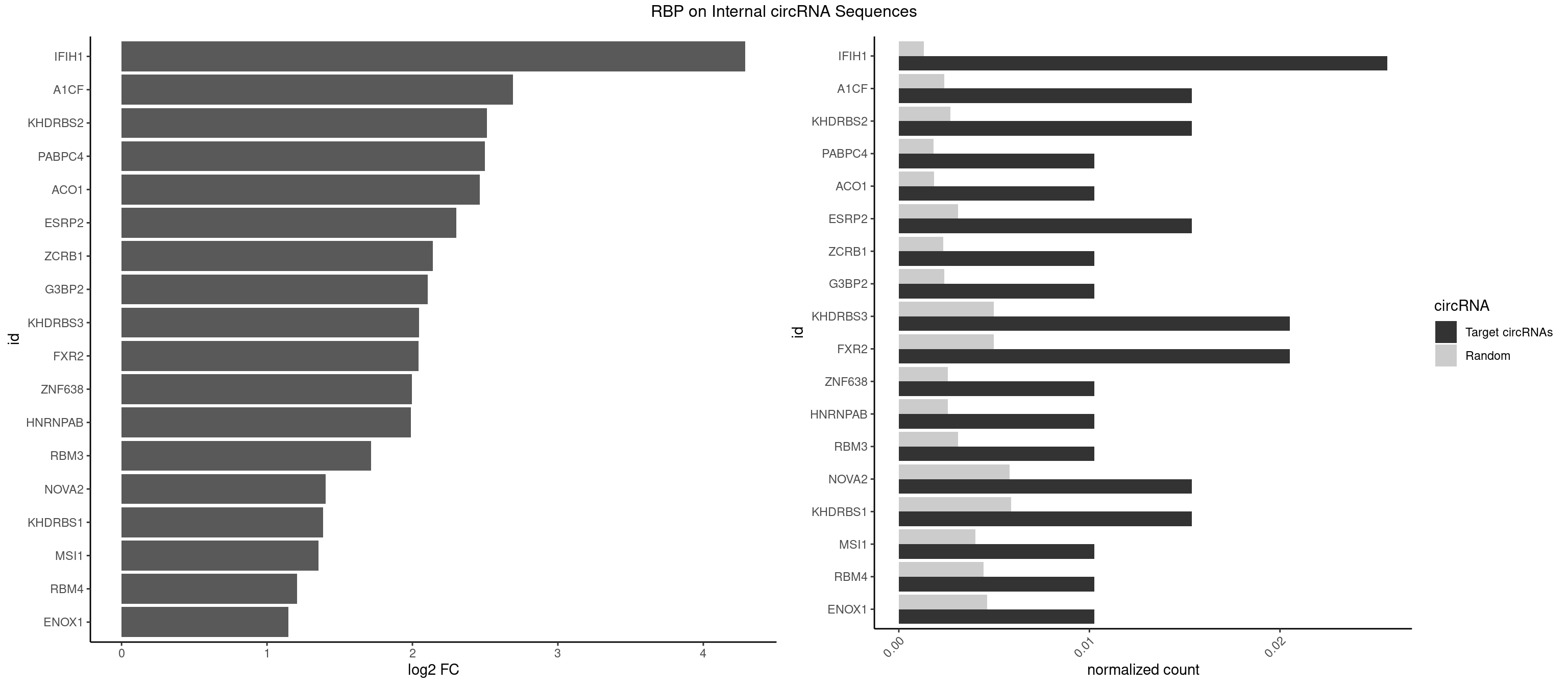
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IFIH1 | 4 | 904 | 0.02564103 | 0.001311530 | 4.289132 | CCGCGG,CGCGGA,GCCGCG,GCGGAU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| A1CF | 2 | 1642 | 0.01538462 | 0.002381042 | 2.691823 | GAUCAG,UCAGUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| KHDRBS2 | 2 | 1858 | 0.01538462 | 0.002694070 | 2.513629 | AUAAAA,GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| PABPC4 | 1 | 1251 | 0.01025641 | 0.001814403 | 2.498959 | AAAAAG | AAAAAA,AAAAAG |
| ACO1 | 1 | 1283 | 0.01025641 | 0.001860778 | 2.462548 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ESRP2 | 2 | 2150 | 0.01538462 | 0.003117238 | 2.303148 | GGGGAU,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| ZCRB1 | 1 | 1605 | 0.01025641 | 0.002327422 | 2.139721 | AAUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| G3BP2 | 1 | 1644 | 0.01025641 | 0.002383941 | 2.105106 | GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| KHDRBS3 | 3 | 3429 | 0.02051282 | 0.004970770 | 2.044985 | AGAUAA,AUAAAA,GAUAAA | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| FXR2 | 3 | 3434 | 0.02051282 | 0.004978016 | 2.042883 | AGACAA,AGACAG,GACAGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| ZNF638 | 1 | 1773 | 0.01025641 | 0.002570888 | 1.996187 | GGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPAB | 1 | 1782 | 0.01025641 | 0.002583931 | 1.988887 | AGACAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 1 | 2152 | 0.01025641 | 0.003120136 | 1.716845 | AAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| NOVA2 | 2 | 4013 | 0.01538462 | 0.005817105 | 1.403115 | GAGACA | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS1 | 2 | 4064 | 0.01538462 | 0.005891014 | 1.384900 | AUAAAA,UAAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| MSI1 | 1 | 2770 | 0.01025641 | 0.004015744 | 1.352787 | AGUAAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| RBM4 | 1 | 3065 | 0.01025641 | 0.004443259 | 1.206836 | CGCGGU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ENOX1 | 1 | 3195 | 0.01025641 | 0.004631656 | 1.146926 | AGACAG | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
RBP on BSJ (Exon Only)
Plot
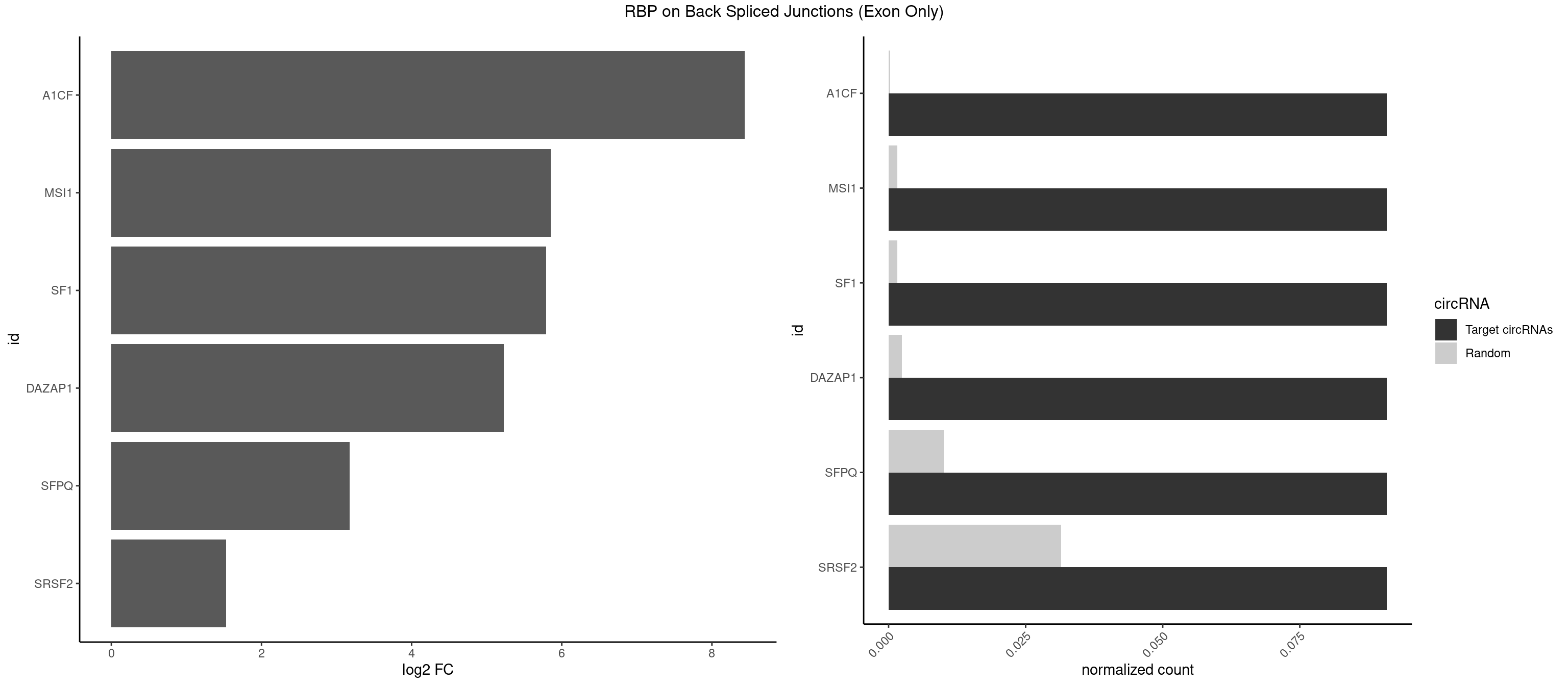
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| A1CF | 1 | 3 | 0.09090909 | 0.0002619859 | 8.438792 | UCAGUA | AUCAGU,CAGUAU,UAAUUG |
| MSI1 | 1 | 23 | 0.09090909 | 0.0015719151 | 5.853829 | AGUAAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUGG,UAGGAA,UAGGAG,UAGGUG,UAGUUA,UAGUUG |
| SF1 | 1 | 24 | 0.09090909 | 0.0016374116 | 5.794936 | AGUAAG | ACAGAC,ACAGUC,AGUAAG,AUACUA,CACAGA,CACUGA,CAGUCA,GCUGAC,GCUGCC,UACUGA,UGCUAA,UGCUGA,UGCUGC |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.0024233691 | 5.229338 | AGUAAG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | GUAAGA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383023 | 1.531901 | GUCAGU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
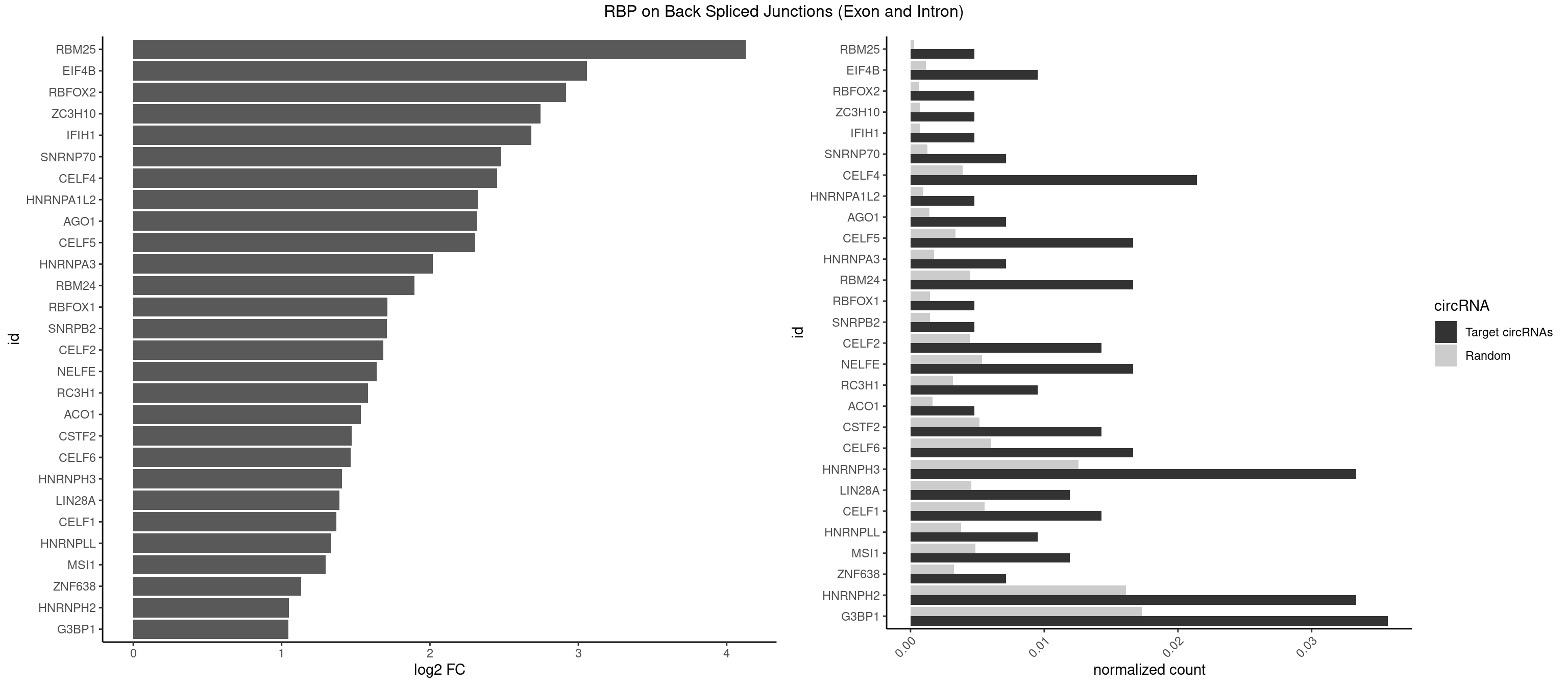
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| EIF4B | 3 | 226 | 0.009523810 | 0.0011436921 | 3.057840 | CUCGGA,GUCGGA,UCGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| ZC3H10 | 1 | 140 | 0.004761905 | 0.0007103990 | 2.744837 | GAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF4 | 8 | 776 | 0.021428571 | 0.0039147521 | 2.452543 | GGUGUG,GUGUGG,GUGUGU,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | GUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | AGGUAG,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| CELF5 | 6 | 669 | 0.016666667 | 0.0033756550 | 2.303726 | GUGUGG,GUGUGU,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AGGAGC,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBM24 | 6 | 888 | 0.016666667 | 0.0044790407 | 1.895704 | AGAGUG,GAGUGA,GUGUGG,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| CELF2 | 5 | 881 | 0.014285714 | 0.0044437727 | 1.684716 | AUGUGU,GUCUGU,GUGUGU,UGUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| NELFE | 6 | 1059 | 0.016666667 | 0.0053405885 | 1.641895 | CUCUGG,CUGGCU,GGUCUC,GUCUCU,UCUGGU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CCCUUC,CUUCUG,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUGU,GUGUUU,UGUGUG,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF6 | 6 | 1196 | 0.016666667 | 0.0060308343 | 1.466536 | GUGAGG,GUGUGG,UGUGAG,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPH3 | 13 | 2496 | 0.033333333 | 0.0125806127 | 1.405763 | AAUGUG,CGAGGG,GGAGGA,GGAGGG,GGGAGG,GGGCGU,GGGCUG,GGGUGU,GUCGAG,UCGAGG,UCGGGC | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | GGAGGA,GGAGGG,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | GUGUGU,UGUGUG,UGUGUU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPLL | 3 | 748 | 0.009523810 | 0.0037736800 | 1.335567 | CACACC,CACUGC,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGAGG,AGGUAG,AGUAAG,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUGG,GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPH2 | 13 | 3198 | 0.033333333 | 0.0161174929 | 1.048338 | AAUGUG,CGAGGG,GGAGGA,GGAGGG,GGGAGG,GGGCGU,GGGCUG,GGGUGU,GUCGAG,UCGAGG,UCGGGC | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| G3BP1 | 14 | 3431 | 0.035714286 | 0.0172914148 | 1.046445 | ACCCCC,AGGCAC,AGGCCC,CAGGCC,CCCCCG,CCCCGG,CCCCUC,CCCUCG,CCGGCC,CCUCGG,CGGCAG,CGGCCG | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.