circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000078304:+:14:101856685:101856885
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000078304:+:14:101856685:101856885 | ENSG00000078304 | ENST00000694906 | + | 14 | 101856686 | 101856885 | 200 | AUGUUCCUCCUGCUGAUCAAGAGAAGCUUUUUAUCCAGAAGUUACGUCAGUGUUGCGUCCUCUUUGACUUUGUUUCUGAUCCACUAAGUGACCUAAAGUGGAAGGAAGUAAAACGAGCUGCUUUAAGUGAAAUGGUAGAAUAUAUCACCCAUAAUCGGAAUGUGAUCACAGAGCCUAUUUACCCAGAAGUAGUCCAUAUGAUGUUCCUCCUGCUGAUCAAGAGAAGCUUUUUAUCCAGAAGUUACGUCAG | circ |
| ENSG00000078304:+:14:101856685:101856885 | ENSG00000078304 | ENST00000694906 | + | 14 | 101856686 | 101856885 | 22 | UAGUCCAUAUGAUGUUCCUCCU | bsj |
| ENSG00000078304:+:14:101856685:101856885 | ENSG00000078304 | ENST00000694906 | + | 14 | 101856486 | 101856695 | 210 | CCAAACAUAUAAUAUGACCAGCGACUAGCUGGGUUUUUUCGUUUUAUUUUCUUUAAGGAAUAGCCUUUGUUUACCCCACCCAUCUCCUUUUUUCUUGAAUCUCUAUUCAAAAUAAAAAUAGAAGAAAGCUUAAAUGUGUUUCACAAUAACUUUGGGUUAAGCACUUUUGUGAUCAAUAUACUUUGCUUUCUGCUUUUCAGAUGUUCCUCC | ie_up |
| ENSG00000078304:+:14:101856685:101856885 | ENSG00000078304 | ENST00000694906 | + | 14 | 101856876 | 101857085 | 210 | AGUCCAUAUGGUAAGUGAUUACAGUUUAACCAGUGUGUCCCAGUUCUGAUUUAUAAACAGGACAGGCCAAGAUCUCUGAGCCUAACACAGUGCUACCCAGGAGAAGAAUCCUCCUGUUCCAGAAUUUGUAGUUGUGCUUGAAGAAUGAAAAUUUUCUUUGUAUCCUGGAUUGUAAUGAUAAUAAUAGGAAGGGAAGUCAGUUGUCUUCUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
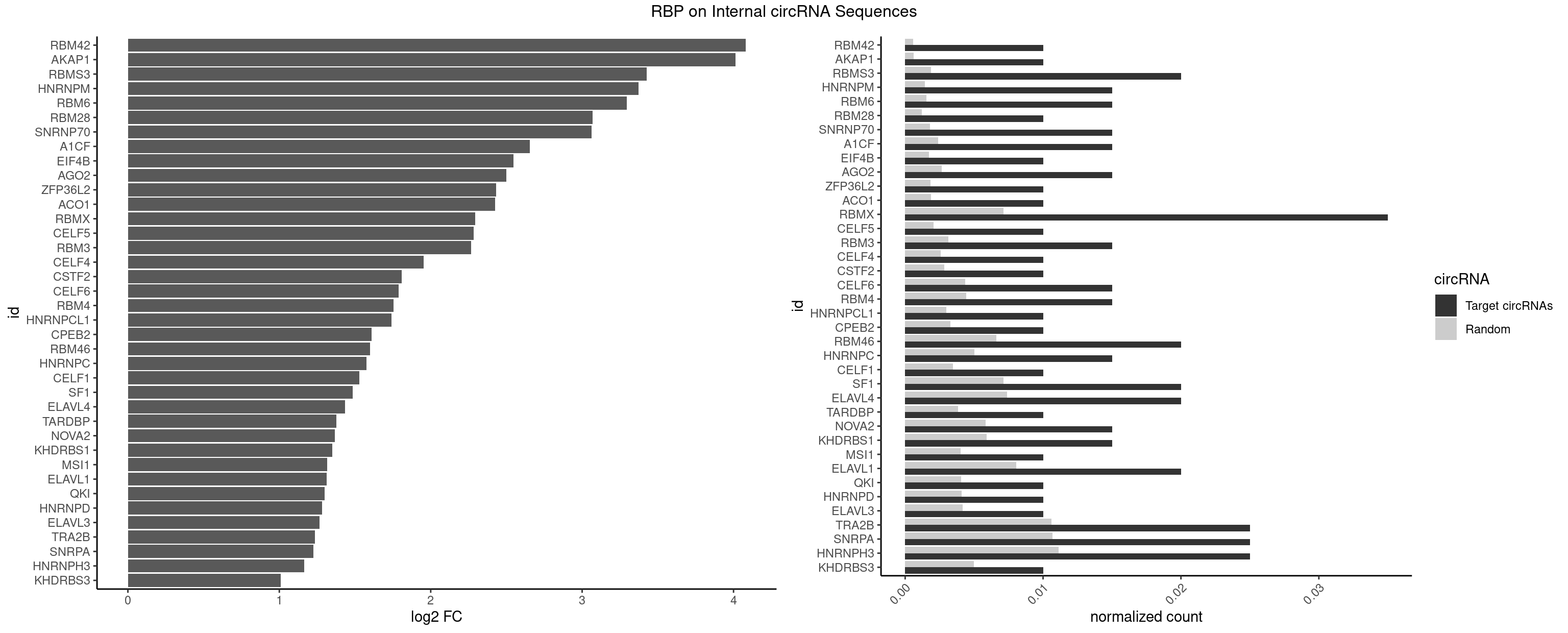
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM42 | 1 | 407 | 0.010 | 0.0005912752 | 4.080026 | ACUAAG | AACUAA,AACUAC,ACUAAG,ACUACG |
| AKAP1 | 1 | 426 | 0.010 | 0.0006188101 | 4.014359 | AUAUAU | AUAUAU,UAUAUA |
| RBMS3 | 3 | 1283 | 0.020 | 0.0018607779 | 3.426022 | AAUAUA,AUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| HNRNPM | 2 | 999 | 0.015 | 0.0014492040 | 3.371630 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM6 | 2 | 1054 | 0.015 | 0.0015289102 | 3.294387 | AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBM28 | 1 | 822 | 0.010 | 0.0011926949 | 3.067703 | AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SNRNP70 | 2 | 1237 | 0.015 | 0.0017941145 | 3.063619 | AUCAAG,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| A1CF | 2 | 1642 | 0.015 | 0.0023810421 | 2.655297 | UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| EIF4B | 1 | 1179 | 0.010 | 0.0017100607 | 2.547881 | UCGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| AGO2 | 2 | 1830 | 0.015 | 0.0026534924 | 2.498998 | AAAGUG,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| ZFP36L2 | 1 | 1277 | 0.010 | 0.0018520827 | 2.432780 | UAUUUA | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| ACO1 | 1 | 1283 | 0.010 | 0.0018607779 | 2.426022 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMX | 6 | 4925 | 0.035 | 0.0071387787 | 2.293606 | AAGGAA,AAGUAA,AGGAAG,AGUGUU,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF5 | 1 | 1415 | 0.010 | 0.0020520728 | 2.284846 | GUGUUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM3 | 2 | 2152 | 0.015 | 0.0031201361 | 2.265282 | AAAACG,AAACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CELF4 | 1 | 1782 | 0.010 | 0.0025839306 | 1.952361 | GUGUUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CSTF2 | 1 | 1967 | 0.010 | 0.0028520334 | 1.809937 | GUGUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF6 | 2 | 2997 | 0.015 | 0.0043447134 | 1.787630 | GUGUUG,UGUGAU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM4 | 2 | 3065 | 0.015 | 0.0044432593 | 1.755272 | CCUCUU,CUCUUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| HNRNPCL1 | 1 | 2062 | 0.010 | 0.0029897078 | 1.741924 | CUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB2 | 1 | 2261 | 0.010 | 0.0032780993 | 1.609069 | CUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| RBM46 | 3 | 4554 | 0.020 | 0.0066011240 | 1.599216 | AUCAAG,AUGAUG,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPC | 2 | 3471 | 0.015 | 0.0050316361 | 1.575863 | CUUUUU,UUUUUA | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CELF1 | 1 | 2391 | 0.010 | 0.0034664959 | 1.528450 | GUGUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| SF1 | 3 | 4931 | 0.020 | 0.0071474739 | 1.484495 | ACUAAG,CACAGA,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| ELAVL4 | 3 | 5105 | 0.020 | 0.0073996354 | 1.434474 | UAUUUA,UUUUAU,UUUUUA | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TARDBP | 1 | 2654 | 0.010 | 0.0038476365 | 1.377956 | GAAUGU | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| NOVA2 | 2 | 4013 | 0.015 | 0.0058171047 | 1.366589 | AUCACC,GAUCAC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| KHDRBS1 | 2 | 4064 | 0.015 | 0.0058910141 | 1.348375 | AUUUAC,UAAAAC | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| MSI1 | 1 | 2770 | 0.010 | 0.0040157442 | 1.316261 | AGGAAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ELAVL1 | 3 | 5554 | 0.020 | 0.0080503280 | 1.312881 | UAUUUA,UUGUUU,UUUGUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| QKI | 1 | 2805 | 0.010 | 0.0040664663 | 1.298152 | CACUAA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| HNRNPD | 1 | 2837 | 0.010 | 0.0041128408 | 1.281793 | UAUUUA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ELAVL3 | 1 | 2867 | 0.010 | 0.0041563169 | 1.266622 | UAUUUA | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| TRA2B | 4 | 7329 | 0.025 | 0.0106226650 | 1.234782 | AAGGAA,AGGAAG,GAAGGA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SNRPA | 4 | 7380 | 0.025 | 0.0106965744 | 1.224779 | AGUAGU,CCUGCU,GUAGUC,UCCUGC | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| HNRNPH3 | 4 | 7688 | 0.025 | 0.0111429292 | 1.165800 | AAUGUG,GAAUGU,GGAAGG,GGAAUG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| KHDRBS3 | 1 | 3429 | 0.010 | 0.0049707696 | 1.008459 | UAAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
RBP on BSJ (Exon Only)
Plot
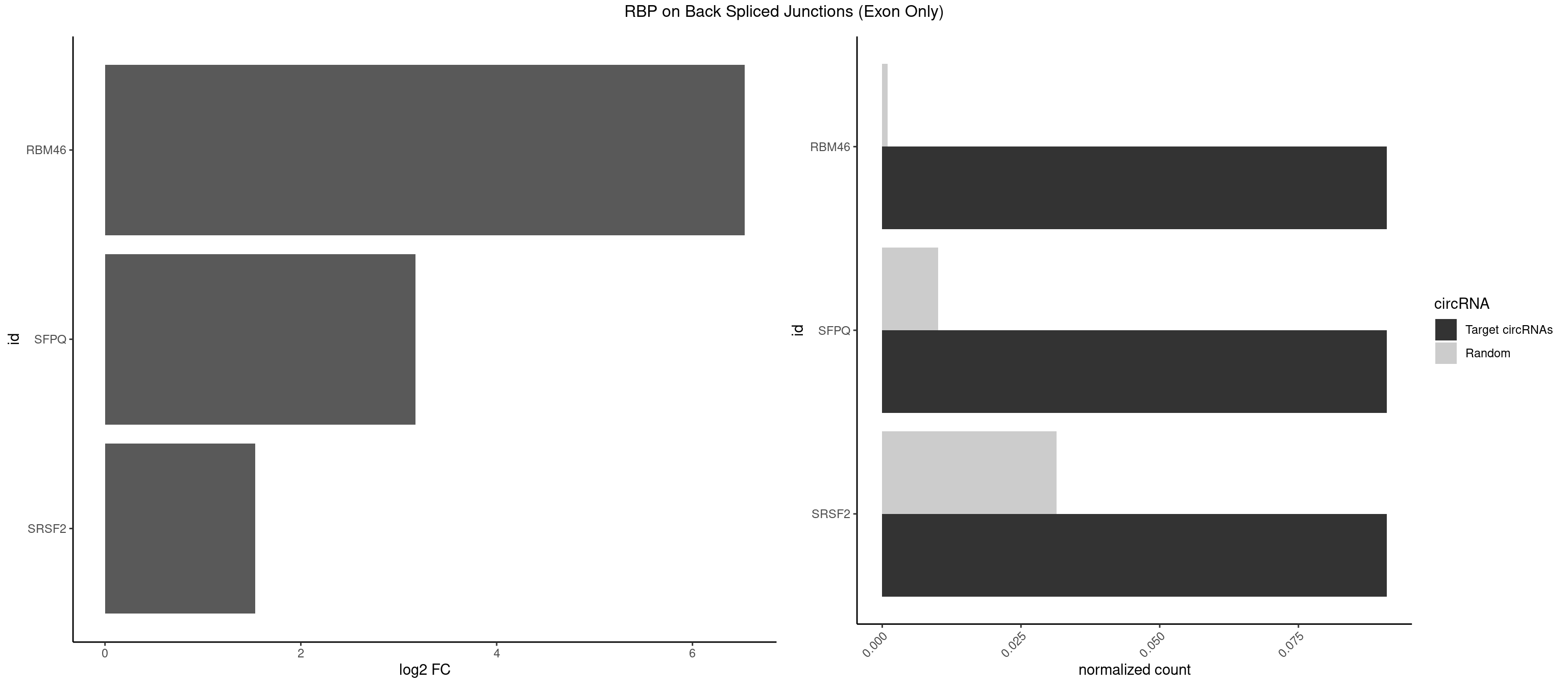
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM46 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | AUGAUG | AAUCAA,AAUGAU,AUCAAA,AUCAAU,AUGAAG,AUGAUG,AUGAUU,GAUCAU,GAUGAA,GAUGAU |
| SFPQ | 1 | 153 | 0.09090909 | 0.0100864553 | 3.172005 | UGAUGU | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| SRSF2 | 1 | 479 | 0.09090909 | 0.0314383023 | 1.531901 | AUGAUG | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
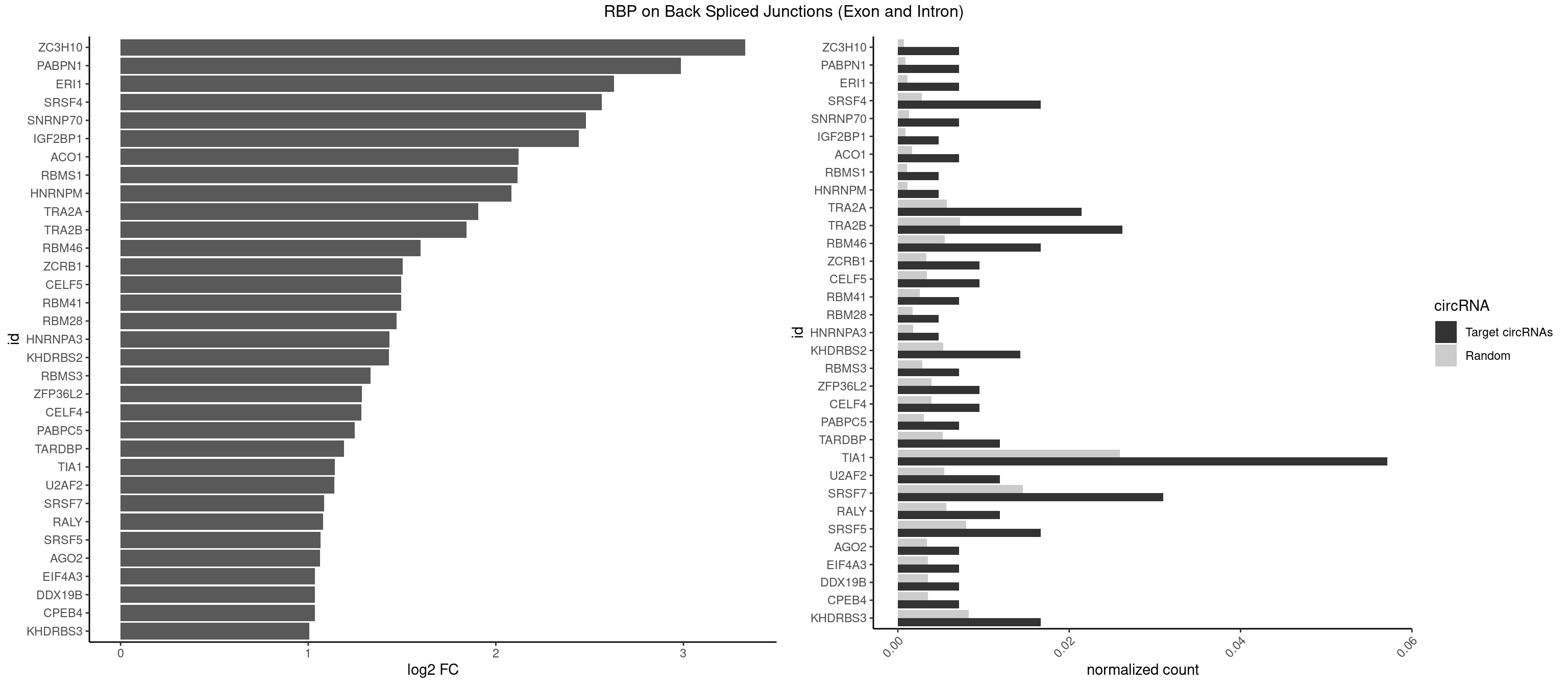
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZC3H10 | 2 | 140 | 0.007142857 | 0.0007103990 | 3.329800 | CAGCGA,CCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AGAAGA | AAAAGA,AGAAGA |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| SRSF4 | 6 | 558 | 0.016666667 | 0.0028164047 | 2.565039 | AGAAGA,AGGAAG,GAAGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AUUCAA,GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AAGCAC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGC,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAC | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| TRA2A | 8 | 1133 | 0.021428571 | 0.0057134220 | 1.907109 | AAGAAA,AGAAAG,AGAAGA,AGGAAG,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| TRA2B | 10 | 1447 | 0.026190476 | 0.0072954454 | 1.843974 | AAGAAU,AAGGAA,AGAAGA,AGGAAG,GAAGAA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AAUGAA,AAUGAU,AUCAAU,AUGAAA,AUGAUA,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | GAUUUA,GCUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGU,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | AUACUU,UACUUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| KHDRBS2 | 5 | 1051 | 0.014285714 | 0.0053002821 | 1.430432 | AAUAAA,AUAAAA,AUAAAC,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | AAUAUA,CAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZFP36L2 | 3 | 774 | 0.009523810 | 0.0039046755 | 1.286336 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CELF4 | 3 | 776 | 0.009523810 | 0.0039147521 | 1.282618 | GUGUGU,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GAAUGA,GUGUGU,GUUGUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| TIA1 | 23 | 5140 | 0.057142857 | 0.0259018541 | 1.141518 | AUUUUC,CUCCUU,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGU,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| SRSF7 | 12 | 2893 | 0.030952381 | 0.0145808142 | 1.085979 | AAUGAU,AGAAGA,AGAUCU,AGGAAG,AGGACA,AUAGAA,GAAGAA,GAAUGA,GGAAGG | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | AGAAGA,AGGAAG,GAAGAA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 0.0082980653 | 1.006119 | AAAUAA,AAUAAA,AUAAAA,AUAAAC,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.