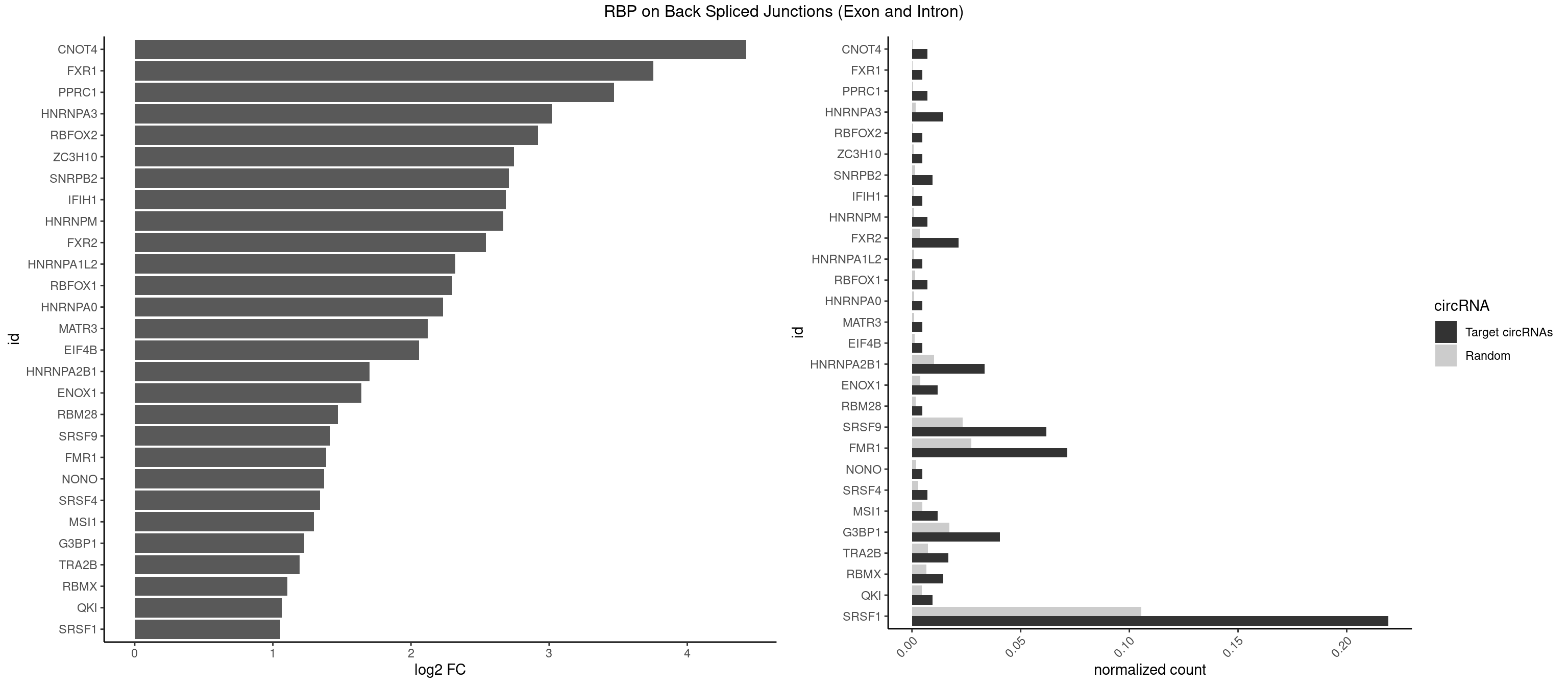
| CNOT4 |
2 |
65 |
0.007142857 |
0.0003325272 |
4.424957 |
GACAGA |
GACAGA |
| FXR1 |
1 |
69 |
0.004761905 |
0.0003526804 |
3.755106 |
AUGACA |
ACGACA,ACGACG,AUGACA,AUGACG |
| PPRC1 |
2 |
127 |
0.007142857 |
0.0006449012 |
3.469351 |
GGCGCC,GGGCGC |
CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| HNRNPA3 |
5 |
349 |
0.014285714 |
0.0017634019 |
3.018140 |
AAGGAG,AGGAGC,GGAGCC |
AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| RBFOX2 |
1 |
124 |
0.004761905 |
0.0006297864 |
2.918604 |
UGCAUG |
UGACUG,UGCAUG |
| ZC3H10 |
1 |
140 |
0.004761905 |
0.0007103990 |
2.744837 |
GGAGCG |
CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| SNRPB2 |
3 |
288 |
0.009523810 |
0.0014560661 |
2.709463 |
AUUGCA,GUAUUG,UUGCAG |
AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| IFIH1 |
1 |
146 |
0.004761905 |
0.0007406288 |
2.684716 |
GGGCCG |
CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| HNRNPM |
2 |
222 |
0.007142857 |
0.0011235389 |
2.668451 |
GAAGGA |
AAGGAA,GAAGGA,GGGGGG |
| FXR2 |
8 |
730 |
0.021428571 |
0.0036829907 |
2.540586 |
GACAGA,GACAGG,GGACAG,UGACAG |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPA1L2 |
1 |
188 |
0.004761905 |
0.0009522370 |
2.322146 |
GUAGGG |
AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBFOX1 |
2 |
287 |
0.007142857 |
0.0014510278 |
2.299426 |
GCAUGA,UGCAUG |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA0 |
1 |
200 |
0.004761905 |
0.0010126965 |
2.233337 |
AGUAGG |
AAUUUA,AGAUAU,AGUAGG |
| MATR3 |
1 |
216 |
0.004761905 |
0.0010933091 |
2.122837 |
AUCUUG |
AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| EIF4B |
1 |
226 |
0.004761905 |
0.0011436921 |
2.057840 |
UUGGAA |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPA2B1 |
13 |
2033 |
0.033333333 |
0.0102478839 |
1.701640 |
AAGGAG,AGGAGC,AGGGGC,AGUAGG,GAAGCC,GAAGGA,GGAGCC,GGGGCC,GUAGGG |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| ENOX1 |
4 |
756 |
0.011904762 |
0.0038139863 |
1.642167 |
CGGACA,GGACAG |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RBM28 |
1 |
340 |
0.004761905 |
0.0017180572 |
1.470761 |
AGUAGG |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SRSF9 |
25 |
4608 |
0.061904762 |
0.0232214833 |
1.414590 |
AGGAAA,AGGAGC,AUGACA,GAAGCC,GAAGGA,GGAAAA,GGAAGG,GGACAG,GGAGCC,GGAGCG,GGGAGG,GGGUGG,GGUGGA,GGUGGC,UGAAAG,UGAAGC,UGACAG,UGGAGC |
AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| FMR1 |
29 |
5430 |
0.071428571 |
0.0273629585 |
1.384277 |
AAGGAG,AGGAAG,AGGCAC,AGGGAG,CUAAGG,CUGGGC,GAAGGA,GACAGG,GAGCGG,GCGACU,GCUAAG,GGACAG,GGCUAA,GGCUGA,GGGCUA,GGGCUG,GUGGCU,UAAGGA,UAGGCA,UGACAG,UGAGGA,UGGCUG |
AAAAAA,AAGCGG,AAGGAA,AAGGAG,AAGGAU,AAGGGA,ACUAAG,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGCGAC,AGCGGC,AGCUGG,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUAGGC,AUGGAG,CAGCUG,CAUAGG,CGAAGG,CGACUG,CGGCUG,CUAAGG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GACUAA,GACUGG,GAGCGA,GAGCGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCGACU,GCGGCU,GCUAAG,GCUAUG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGACUA,GGCAUA,GGCGAA,GGCUAA,GGCUAU,GGCUGA,GGCUGG,GGGCAU,GGGCGA,GGGCUA,GGGCUG,GGGGGG,GUGCGA,GUGCGG,GUGGCU,UAAGGA,UAGCAG,UAGCGA,UAGCGG,UAGGCA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGAC,UGCGGC,UGGAGU,UGGCUG,UUUUUU |
| NONO |
1 |
364 |
0.004761905 |
0.0018389762 |
1.372636 |
GAGGAA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SRSF4 |
2 |
558 |
0.007142857 |
0.0028164047 |
1.342647 |
AGGAAG,GAGGAA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| MSI1 |
4 |
961 |
0.011904762 |
0.0048468360 |
1.296424 |
AGGAAG,AGGUGG,AGUAGG,UAGUUA |
AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| G3BP1 |
16 |
3431 |
0.040476190 |
0.0172914148 |
1.227018 |
ACAGGC,ACCCCU,AGGCAC,CACAGG,CCACCC,CCAGGC,CCCAGG,CCCCAG,CCCCGC,CCCCUC,CCCGCC,CCCUCC,UAGGCA |
ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| TRA2B |
6 |
1447 |
0.016666667 |
0.0072954454 |
1.191898 |
AGGAAA,AGGAAG,GAAGGA,GGAAGG |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBMX |
5 |
1316 |
0.014285714 |
0.0066354293 |
1.106311 |
AGGAAG,GAAGGA,GGAAGG |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| QKI |
3 |
904 |
0.009523810 |
0.0045596534 |
1.062615 |
ACUCAU,CUCAUA,UCAUAU |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SRSF1 |
91 |
20925 |
0.219047619 |
0.1054312777 |
1.054942 |
AAGGAC,AAGGAG,ACAGAG,ACAGGG,ACAGGU,ACCCGG,AGAGGG,AGCGGU,AGGAAA,AGGAAG,AGGACU,AGGAGC,AGGCGA,AUGAAG,AUGACA,CACAGG,CAGAGG,CCACCC,CCAGCA,CCAGGG,CCCAGG,CCCCGC,CCCGCC,CCCGGG,CCCUCC,CCCUGC,CCCUGG,CCGGAC,CCGGGC,CCUCCA,CCUGCA,CCUGGG,CGACCC,CGCCCC,CGGACA,CGGGCC,CGGUGG,CUCAGG,CUGAAC,GAAGCC,GAAGGA,GACAGA,GACAGG,GACCCG,GACUGA,GAGCGG,GAGGAA,GAGGCA,GAGGGC,GAUGAA,GCACGU,GCAGGG,GCCGGA,GCGCCC,GCUCCA,GGAAGG,GGACAG,GGACUG,GGAGCC,GGAGCG,GGAGCU,GGAGGG,GGAGGU,GGCACA,GGCACG,GGCCGG,GGGCCA,GGGCCG,GGGGAG,GGUGGA,GGUGGC,UGAACC,UGAAGC,UGACAG,UGAUGA,UGGAGC |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AACACG,AACAGC,AACCGG,AAGAAC,AAGAAG,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGAG,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACACGA,ACACGU,ACAGAA,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCCGG,ACCCGU,ACCGGA,ACCGGU,ACGAAC,ACGAAG,ACGAAU,ACGACG,ACGCGA,ACGCGC,ACGCUC,ACGGAA,ACGGAC,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGA,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCCGU,AGCGAG,AGCGGA,AGCGGG,AGCGGU,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGCGA,AGGUAA,AGUCGG,AUACGA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,AUUACG,AUUUCA,CAACCA,CAACCC,CAACCG,CAACGA,CAACGC,CAAGCA,CAAGGA,CAAUGG,CACACG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGA,CACCGG,CACGCA,CACGCU,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGCGG,CAGUCG,CAUGGU,CCAACC,CCAACG,CCAAGC,CCAAGG,CCAAUG,CCACAG,CCACCA,CCACCC,CCACCG,CCACGA,CCACGC,CCACGG,CCACUG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCAUGA,CCAUGG,CCCACC,CCCACG,CCCACU,CCCAGC,CCCAGG,CCCAUG,CCCCCA,CCCCCC,CCCCCG,CCCCGA,CCCCGC,CCCCGG,CCCGAC,CCCGCA,CCCGCC,CCCGCG,CCCGGA,CCCGGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCCUGA,CCCUGC,CCCUGG,CCGACA,CCGACC,CCGACG,CCGACU,CCGAGC,CCGAGG,CCGAUG,CCGCCA,CCGCCC,CCGCCG,CCGCGA,CCGCGC,CCGCGG,CCGCUA,CCGGAC,CCGGAG,CCGGCA,CCGGCC,CCGGCG,CCGGGA,CCGGGC,CCGGGG,CCGUCC,CCGUCG,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCA,CCUCCG,CCUCGA,CCUCGG,CCUGAC,CCUGCA,CCUGCG,CCUGGA,CCUGGG,CGAACC,CGAACG,CGAAGC,CGAAGG,CGACAG,CGACCA,CGACCC,CGACCG,CGACGA,CGACGC,CGACGG,CGAGCA,CGAGCG,CGAGGA,CGAGGC,CGAGGG,CGAUGA,CGAUGG,CGCACA,CGCACC,CGCACG,CGCAGC,CGCAGG,CGCCCA,CGCCCC,CGCCCG,CGCCGA,CGCCGC,CGCCGG,CGCCGU,CGCGAG,CGCGCA,CGCGCC,CGCGCG,CGCGCU,CGCGGA,CGCGGC,CGCGGG,CGCGGU,CGCGUC,CGCUAU,CGCUCA,CGCUCC,CGCUCG,CGCUGC,CGCUGG,CGGAAC,CGGAAG,CGGAAU,CGGACA,CGGACC,CGGACG,CGGAGC,CGGAGG,CGGCAC,CGGCAG,CGGCCA,CGGCCC,CGGCCG,CGGCGA,CGGCGC,CGGCGG,CGGGCA,CGGGCC,CGGGCG,CGGGGA,CGGGGC,CGGGGG,CGGUCC,CGGUCG,CGGUGC,CGGUGG,CGUCCA,CGUCCG,CGUCGG,CGUGCA,CGUGCG,CGUGGA,CGUGGG,CUACGA,CUAGGG,CUCAGG,CUCCGG,CUCGGA,CUCGUG,CUGAAC,CUGACU,CUGAGC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAACCA,GAACGA,GAAGAA,GAAGAC,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACCCG,GACCGA,GACGAA,GACGAC,GACGAG,GACGCA,GACGCG,GACGGA,GACGGC,GACGGG,GACUGA,GACUGG,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGCGA,GAGCGG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUACG,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GAUGGU,GCACCA,GCACCG,GCACGA,GCACGG,GCACGU,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCAUAC,GCCCAC,GCCCCA,GCCCCG,GCCCGA,GCCCGG,GCCCGU,GCCGCA,GCCGCG,GCCGGA,GCCGGG,GCCGGU,GCCGUA,GCGAGC,GCGCAA,GCGCCA,GCGCCC,GCGCCG,GCGCGA,GCGCGC,GCGCGG,GCGGAA,GCGGAC,GCGGAG,GCGGCA,GCGGCG,GCGGGA,GCGGGG,GCGGUU,GCGUCA,GCUCCA,GCUCCG,GCUCGA,GCUCGG,GCUGCA,GCUGCG,GCUGGA,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGACGG,GGACGU,GGACUG,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCC,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAC,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCACG,GGCAGA,GGCCCA,GGCCCG,GGCCGA,GGCCGG,GGCCGU,GGCGCA,GGCGCG,GGCGGA,GGCGGG,GGCGGU,GGGACG,GGGCCA,GGGCCG,GGGCGA,GGGCGG,GGGGAA,GGGGAC,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGGGG,GGGUAC,GGUAAC,GGUCCA,GGUCCG,GGUCGA,GGUCGG,GGUGAA,GGUGAC,GGUGAU,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,GUGACG,UAAUUU,UACGAA,UACGGA,UAGACA,UAGGAC,UAUUAC,UCAAGA,UCAGGU,UCCGGA,UCGCAC,UCGGGC,UCGUGU,UGAACA,UGAACC,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUA,UGACUG,UGAGUU,UGAUGA,UGAUGC,UGAUGG,UGGAAA,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGA,UGGUGC,UGGUGG,UGUAGG,UUAAUU,UUACGG,UUCAAG,UUGACA,UUUCAA |