circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000184903:-:7:111487237:111487341
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000184903:-:7:111487237:111487341 | ENSG00000184903 | ENST00000405709 | - | 7 | 111487238 | 111487341 | 104 | CCUUCUUUGAAUCCUGGGGGGAGCCAGUCAUCUGAUGUGGUGCUUUUGAACCACUGGAAAGUGAGGAAUUUUGAAGUACACCGUGGUGACAUUGUAUCAUUGGUCCUUCUUUGAAUCCUGGGGGGAGCCAGUCAUCUGAUGUGGUGCUUUUGAA | circ |
| ENSG00000184903:-:7:111487237:111487341 | ENSG00000184903 | ENST00000405709 | - | 7 | 111487238 | 111487341 | 22 | GUAUCAUUGGUCCUUCUUUGAA | bsj |
| ENSG00000184903:-:7:111487237:111487341 | ENSG00000184903 | ENST00000405709 | - | 7 | 111487332 | 111487541 | 210 | UUUUUUUCUGGUUCAUCUGUGAAGAAUCCCAGUGAGCCUUUUUGAAAACUCAUCAUUUGCAGUUACUUUAGAACAGCAAUUUAUAGAAAGGUACUGUAUUAGAAAUGUUUUAUUUCAGUCCACUCGAGCUGUGAAGUGCAAGAACCAUGAAGAUUGAAAAAUACAAAUGAUAGAAGUGUCUCUUUCUCCUUAUUUUCUAGCCUUCUUUGA | ie_up |
| ENSG00000184903:-:7:111487237:111487341 | ENSG00000184903 | ENST00000405709 | - | 7 | 111487038 | 111487247 | 210 | UAUCAUUGGUGUAAGUAACUCAGAAGCAUACUAUAUUUGUAUAUAAAAUUAUUUAUACUUAUGCUGAUUUAACUGGUAAUCCACUGUUAAUAUCAUAUUGCCAAUUAUUAAAUUGAAUACAUGUUAAAUACAAUGCAUUUUUAAAAAAUCAAUUUUGCAGUGUAACAAUGCUAUAAAAAGAAUUUUUUGCAUUUAAAAUAUGUUAGUUGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
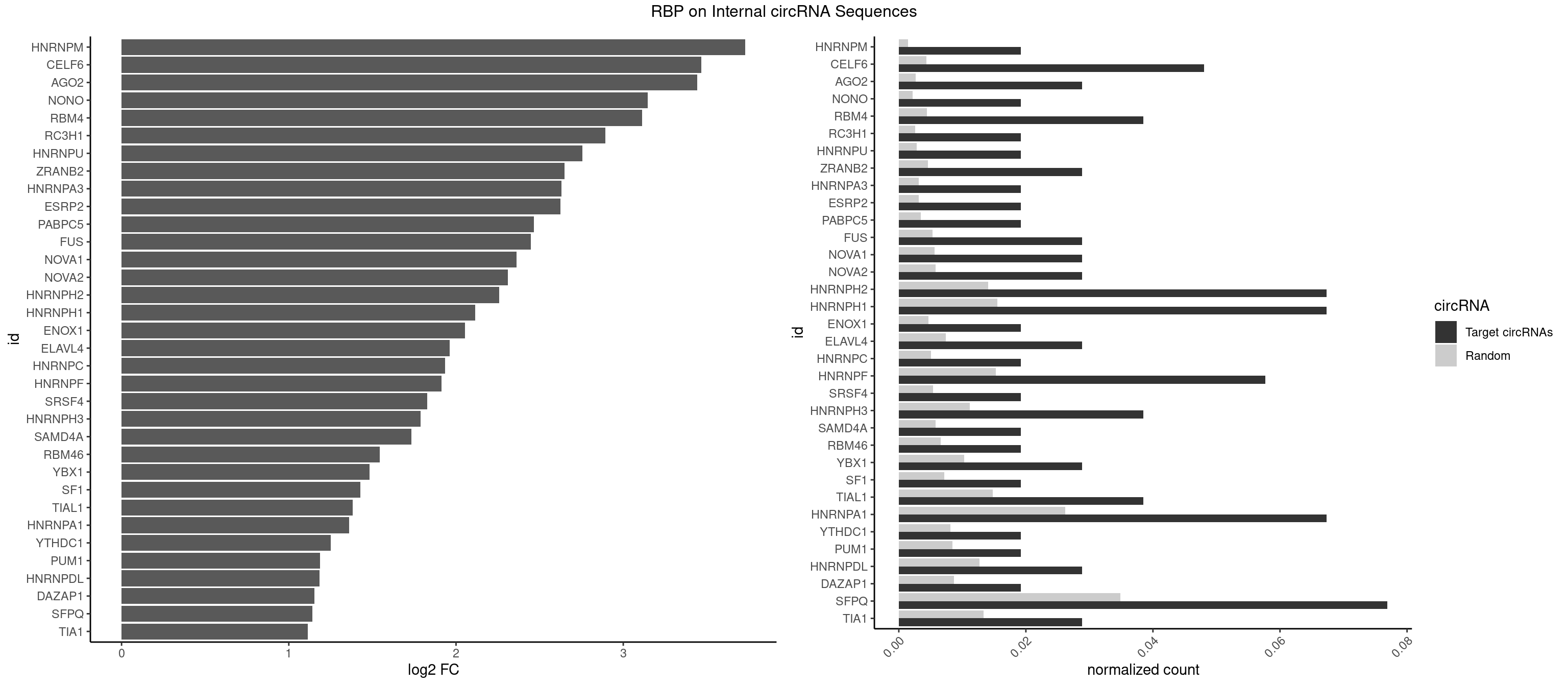
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPM | 1 | 999 | 0.01923077 | 0.001449204 | 3.730084 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| CELF6 | 4 | 2997 | 0.04807692 | 0.004344713 | 3.468012 | GUGAGG,GUGGUG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| AGO2 | 2 | 1830 | 0.02884615 | 0.002653492 | 3.442415 | AAAGUG,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| NONO | 1 | 1498 | 0.01923077 | 0.002172357 | 3.146084 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM4 | 3 | 3065 | 0.03846154 | 0.004443259 | 3.113726 | CCUUCU,CUUCUU,UCCUUC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| RC3H1 | 1 | 1786 | 0.01923077 | 0.002589727 | 2.892544 | CCUUCU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPU | 1 | 1965 | 0.01923077 | 0.002849135 | 2.754821 | GGGGGG | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| ZRANB2 | 2 | 3173 | 0.02884615 | 0.004599773 | 2.648744 | GUGGUG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| HNRNPA3 | 1 | 2140 | 0.01923077 | 0.003102746 | 2.631799 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 1 | 2150 | 0.01923077 | 0.003117238 | 2.625076 | GGGGAG | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| PABPC5 | 1 | 2400 | 0.01923077 | 0.003479539 | 2.466449 | GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| FUS | 2 | 3648 | 0.02884615 | 0.005288145 | 2.447545 | GGGGGG,UGGUGA | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| NOVA1 | 2 | 3872 | 0.02884615 | 0.005612767 | 2.361595 | CAGUCA,GGGGGG | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| NOVA2 | 2 | 4013 | 0.02884615 | 0.005817105 | 2.310006 | AGUCAU,GGGGGG | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| HNRNPH2 | 6 | 9711 | 0.06730769 | 0.014074669 | 2.257670 | AUGUGG,CUGGGG,GGGGAG,GGGGGA,GGGGGG,UGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 6 | 10717 | 0.06730769 | 0.015532568 | 2.115475 | AUGUGG,CUGGGG,GAGGAA,GGGGAG,GGGGGG,UGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| ENOX1 | 1 | 3195 | 0.01923077 | 0.004631656 | 2.053817 | AGUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ELAVL4 | 2 | 5105 | 0.02884615 | 0.007399635 | 1.962853 | UGUAUC,UUGUAU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPC | 1 | 3471 | 0.01923077 | 0.005031636 | 1.934317 | GGGGGG | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPF | 5 | 10561 | 0.05769231 | 0.015306492 | 1.914235 | AUGUGG,CUGGGG,GAGGAA,GGGGAG,GGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| SRSF4 | 1 | 3740 | 0.01923077 | 0.005421472 | 1.826660 | GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPH3 | 3 | 7688 | 0.03846154 | 0.011142929 | 1.787288 | AUGUGG,GGGGAG,GGGGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| SAMD4A | 1 | 3992 | 0.01923077 | 0.005786671 | 1.732611 | CUGGAA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| RBM46 | 1 | 4554 | 0.01923077 | 0.006601124 | 1.542633 | AUCAUU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| YBX1 | 2 | 7119 | 0.02884615 | 0.010318332 | 1.483169 | AACCAC,CAUCUG | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| SF1 | 1 | 4931 | 0.01923077 | 0.007147474 | 1.427911 | CAGUCA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| TIAL1 | 3 | 10182 | 0.03846154 | 0.014757244 | 1.381993 | AAUUUU,AUUUUG,CUUUUG | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPA1 | 6 | 18070 | 0.06730769 | 0.026188565 | 1.361834 | AAGUAC,GAGGAA,GGAGCC,GUGGUG,UGGUGC | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| YTHDC1 | 1 | 5576 | 0.01923077 | 0.008082210 | 1.250595 | UGGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| PUM1 | 1 | 5823 | 0.01923077 | 0.008440164 | 1.188074 | AUUGUA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPDL | 2 | 8759 | 0.02884615 | 0.012695027 | 1.184116 | AUCUGA | AACAGC,AACCUU,AACUAA,AAUACC,AAUUUA,ACACCA,ACAGCA,ACAUUA,ACCACG,ACCAGA,ACCUUG,ACUAAC,ACUAAG,ACUAGA,ACUAGC,ACUAGG,ACUGCA,ACUUUA,AGAUAU,AGUAGC,AGUAGG,AUACCA,AUCUGA,AUGCGC,AUUAGC,AUUAGG,CACCAG,CACGCA,CACUAG,CAUUAG,CCACGC,CCAGAC,CCUUGC,CCUUUA,CGAGCA,CUAACA,CUAACU,CUAAGC,CUAAGU,CUAGAG,CUAGAU,CUAGCA,CUAGCC,CUAGCG,CUAGCU,CUAGGA,CUAGGC,CUAGUA,CUUGCC,CUUUAA,CUUUAG,GAACUA,GACUAG,GAGUAG,GAUUAG,GCACUA,GCGAGC,GCUAGU,GGACUA,GGAGUA,GGAUUA,GUAGCC,GUAGCU,UAACAG,UAAGUA,UAGGCA,UCUGAC,UGCGCA,UGUCGC,UUAGCC,UUAGGC,UUUAGG |
| DAZAP1 | 1 | 5964 | 0.01923077 | 0.008644502 | 1.153562 | GGGGGG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| SFPQ | 7 | 24050 | 0.07692308 | 0.034854804 | 1.142059 | GAGGAA,GGGGGA,GGGGGG,GUGGUG,UGAUGU,UUGAAG | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| TIA1 | 2 | 9206 | 0.02884615 | 0.013342821 | 1.112315 | AUUUUG,CUUUUG | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
RBP on BSJ (Exon Only)
Plot
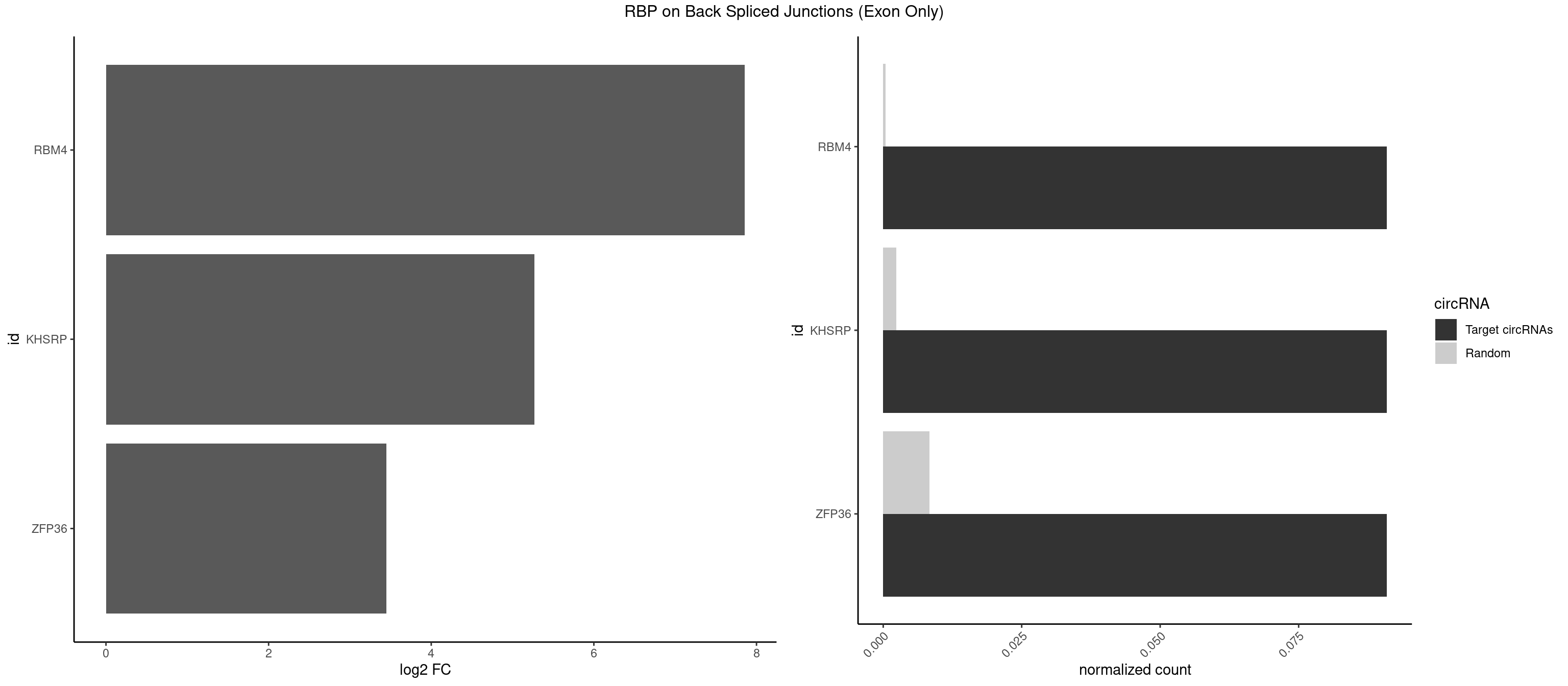
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM4 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | UCCUUC | CCUCUU,CCUUCU,CUCUUU,GCGCGG,UUCUUG |
| KHSRP | 1 | 35 | 0.09090909 | 0.0023578727 | 5.268867 | UCCUUC | ACCCUC,AGCCUC,AUAUUU,AUGUAU,AUGUGU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CGGCUU,CUCCCU,CUGCCU,CUGCUU,GCCCUC,GCCUCC,GCCUUC,GGCCUC,UAGGUU,UAUAUU,UAUUUU,UCCCUC,UGCAUG,UGUAUA,UGUGUG,UUAUUA |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | UCCUUC | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
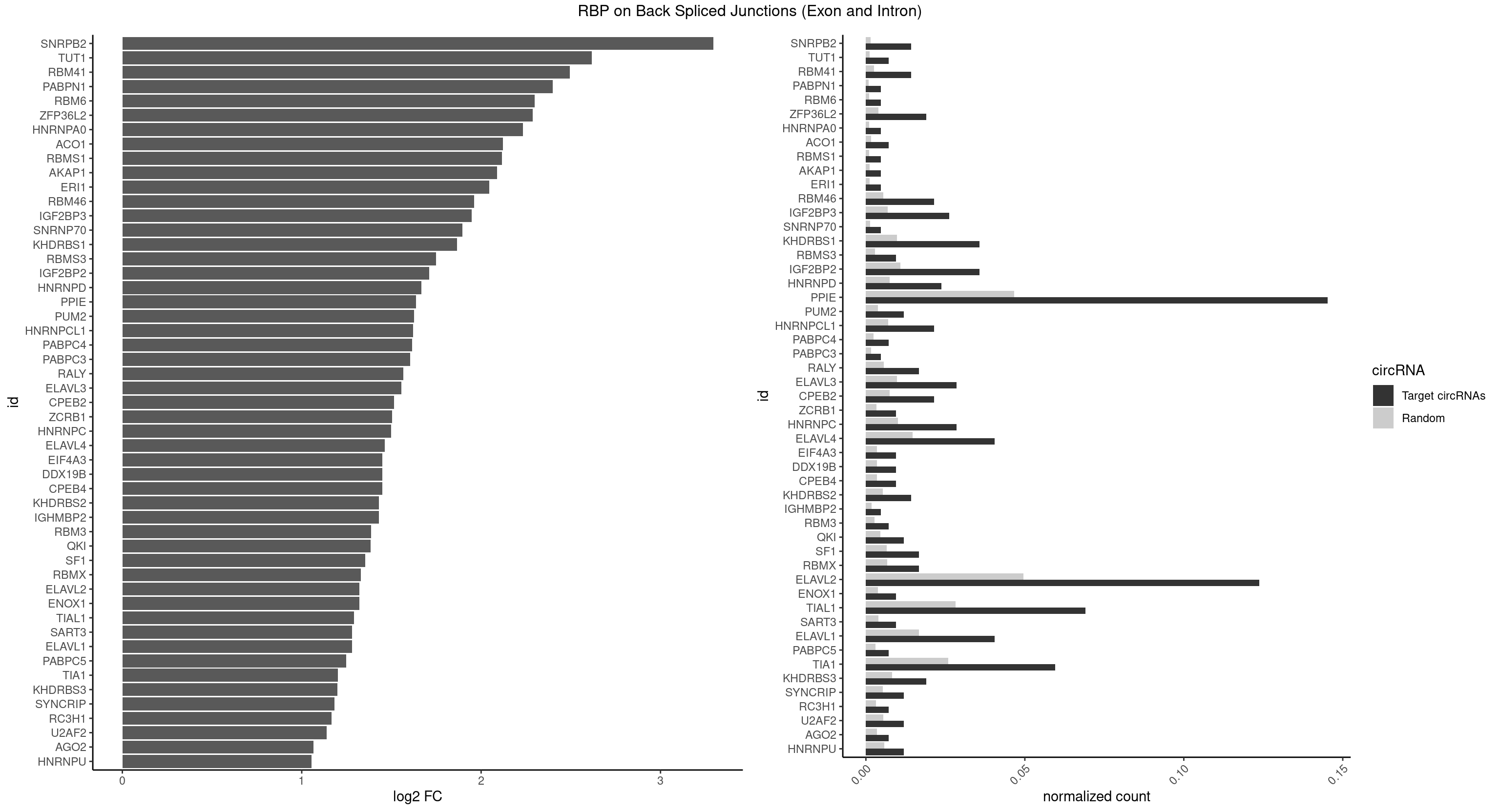
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRPB2 | 5 | 288 | 0.014285714 | 0.0014560661 | 3.294425 | UAUUGC,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM41 | 5 | 502 | 0.014285714 | 0.0025342604 | 2.494937 | AUACAU,AUACUU,UACAUG,UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| RBM6 | 1 | 191 | 0.004761905 | 0.0009673519 | 2.299426 | AAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ZFP36L2 | 7 | 774 | 0.019047619 | 0.0039046755 | 2.286336 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | UAUAUA | AUAUAU,UAUAUA |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| RBM46 | 8 | 1091 | 0.021428571 | 0.0055018138 | 1.961556 | AAUCAA,AAUGAU,AUCAAU,AUCAUA,AUCAUU,AUGAAG,AUGAUA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| IGF2BP3 | 10 | 1348 | 0.026190476 | 0.0067966546 | 1.946146 | AAAAUC,AAACUC,AAAUAC,AAAUCA,AACUCA,AAUACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| KHDRBS1 | 14 | 1945 | 0.035714286 | 0.0098045143 | 1.864983 | AUAAAA,AUUUAA,GAAAAC,UAAAAA,UUAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | CUAUAU,UAUAGA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| IGF2BP2 | 14 | 2163 | 0.035714286 | 0.0109028617 | 1.711794 | AAAAUC,AAACUC,AAAUAC,AAAUCA,AACUCA,AAUACA,CCACUC,GAAAAC,GAAUAC,GCAUAC | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAAAC,CAAACA,CAAAUC,CAACAC,CAACUC,CAAUAC,CAAUCA,CAAUUC,CACACA,CACUCA,CAUACA,CAUUCA,CCAAAC,CCAAUC,CCACAC,CCACUC,CCAUAC,CCAUUC,GAAAAC,GAAAUC,GAACAC,GAACUC,GAAUAC,GAAUUC,GCAAAC,GCAAUC,GCACAC,GCACUC,GCAUAC,GCAUUC |
| HNRNPD | 9 | 1488 | 0.023809524 | 0.0075020153 | 1.666189 | AAAAAA,AAUUUA,AUUUAU,UAUUUA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| PPIE | 60 | 9262 | 0.145238095 | 0.0466696896 | 1.637862 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAU,AAAUUA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAUAA,AUAUUU,AUUAAA,AUUAUU,AUUUAA,AUUUAU,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAU,UAUAAA,UAUAUA,UAUAUU,UAUUAA,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAUUA,UUAUUU,UUUAAA,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUAUAU,UAAAUA,UAUAUA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPCL1 | 8 | 1381 | 0.021428571 | 0.0069629182 | 1.621772 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| RALY | 6 | 1117 | 0.016666667 | 0.0056328094 | 1.565039 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| ELAVL3 | 11 | 1928 | 0.028571429 | 0.0097188634 | 1.555714 | AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| CPEB2 | 8 | 1487 | 0.021428571 | 0.0074969770 | 1.515155 | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AUUUAA,GAUUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPC | 11 | 2006 | 0.028571429 | 0.0101118501 | 1.498526 | AUUUUU,CUUUUU,GCAUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL4 | 16 | 2916 | 0.040476190 | 0.0146966949 | 1.461582 | AAAAAA,AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| CPEB4 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| DDX19B | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 691 | 0.009523810 | 0.0034864974 | 1.449760 | UUUUUU | UUUUUU |
| KHDRBS2 | 5 | 1051 | 0.014285714 | 0.0053002821 | 1.430432 | AUAAAA,UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,AUACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACUCAU,ACUUAU,AUCAUA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SF1 | 6 | 1294 | 0.016666667 | 0.0065245869 | 1.353007 | ACUUAU,AGUAAC,AUACUA,UAACAA,UAUACU,UGCUGA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| RBMX | 6 | 1316 | 0.016666667 | 0.0066354293 | 1.328704 | AAGUAA,AAGUGU,AGUAAC,AUCCCA,GUAACA,UAACAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ELAVL2 | 51 | 9826 | 0.123809524 | 0.0495112858 | 1.322293 | AAUUUA,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAU,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUUCU,CUUUUU,GAUUUA,GUUUUA,UACUUU,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UGAUUU,UGUAUU,UUACUU,UUAGUU,UUAUAC,UUAUUU,UUCUUU,UUGUAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| TIAL1 | 28 | 5597 | 0.069047619 | 0.0282043531 | 1.291674 | AAUUUU,AUUUUC,AUUUUG,AUUUUU,CUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AAAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| ELAVL1 | 16 | 3309 | 0.040476190 | 0.0166767432 | 1.279236 | AUUUAU,UAUUUA,UAUUUU,UGAUUU,UGUUUU,UUAGUU,UUAUUU,UUUAUU,UUUUUG,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| PABPC5 | 2 | 596 | 0.007142857 | 0.0030078597 | 1.247764 | AGAAAG,AGAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| TIA1 | 24 | 5140 | 0.059523810 | 0.0259018541 | 1.200411 | AUUUUC,AUUUUG,AUUUUU,CUCCUU,CUUUUU,GUUUUA,UAUUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCU,UUUUUA,UUUUUC,UUUUUG,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| KHDRBS3 | 7 | 1646 | 0.019047619 | 0.0082980653 | 1.198764 | AUAAAA,AUUAAA,UAAAUA,UUUUUU | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| SYNCRIP | 4 | 1041 | 0.011904762 | 0.0052498992 | 1.181177 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCUUCU,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| AGO2 | 2 | 677 | 0.007142857 | 0.0034159613 | 1.064210 | AAAAAA,AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPU | 4 | 1136 | 0.011904762 | 0.0057285369 | 1.055300 | AAAAAA,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.