circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000197312:+:1:15638306:15643650
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000197312:+:1:15638306:15643650 | ENSG00000197312 | ENST00000480945 | + | 1 | 15638307 | 15643650 | 257 | GCAACAGAACAUUGAGGAAAACAUGACAAUAGCUAUGGAAGAGGCUCCGGAAAGUUUUGGCCAAGUAGUGAUGCUUUAUAUUAACUGCAAAGUGAAUGGACAUCCUGUGAAAGCCUUUGUUGACUCAGGUGCCCAGAUGACUAUCAUGAGCCAAGCUUGUGCAGAAAGGUGUAACAUAAUGAGACUGGUGGACCGUCGGUGGGCAGGGAUUGCCAAAGGAGUGGGCACCCAGAAGAUUAUUGGAAGGGUACAUCUAGGCAACAGAACAUUGAGGAAAACAUGACAAUAGCUAUGGAAGAGGCUCCGG | circ |
| ENSG00000197312:+:1:15638306:15643650 | ENSG00000197312 | ENST00000480945 | + | 1 | 15638307 | 15643650 | 22 | GGUACAUCUAGGCAACAGAACA | bsj |
| ENSG00000197312:+:1:15638306:15643650 | ENSG00000197312 | ENST00000480945 | + | 1 | 15638107 | 15638316 | 210 | AGAUUUUCUGCCAGCUGUUCAUCUUACUGCCUCUUUGGAAAUGAUCUCUUGCCCCAUAUAGCACCAAUUUUUCUAUGACUCGGCUGCUGUGCUUGGGGCAAAAGAUAUGUACAAAUCUGUUUUUUAAUAUUGUGACUUUGAAACUGAUUUCUCUCCAUGGAAUUAAUGCUUUCAGUGUCCCUGGUCUUGUUCUGUUACAGGCAACAGAAC | ie_up |
| ENSG00000197312:+:1:15638306:15643650 | ENSG00000197312 | ENST00000480945 | + | 1 | 15643641 | 15643850 | 210 | GUACAUCUAGGUGAGCAAAAGGCACUGGGGCUUGCUGUCUUUCUGUAGGCUAUUUGUAGGUAGGGUAGUCGAUUUUCAAGUUUUUAGAGGGUCUUUUGUUGUUAUUUUGUUGUUGUUGUUUUAAAUUUCUUUUGUGUGGGUAUGUCUGAGCUAGCAAGCCUAAUUUCUAGAAGUUUUAAAUCCUGUGUUCCGUCAAAGUUAUGUGUUUCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
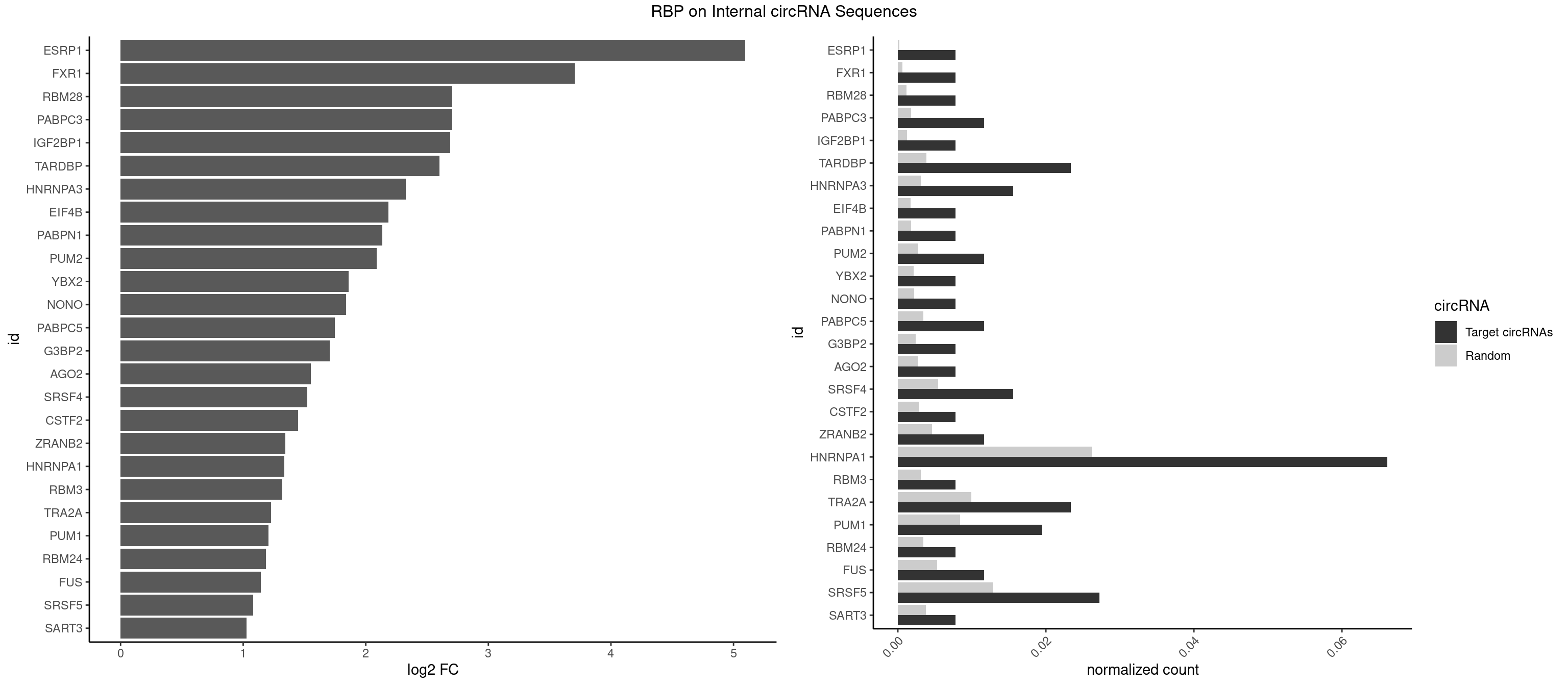
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.007782101 | 0.000227525 | 5.096063 | AGGGAU | AGGGAU |
| FXR1 | 1 | 411 | 0.007782101 | 0.000597072 | 3.704183 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| RBM28 | 1 | 822 | 0.007782101 | 0.001192695 | 2.705935 | AGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| PABPC3 | 2 | 1234 | 0.011673152 | 0.001789767 | 2.705351 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| IGF2BP1 | 1 | 831 | 0.007782101 | 0.001205738 | 2.690244 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| TARDBP | 5 | 2654 | 0.023346304 | 0.003847636 | 2.601150 | GAAUGG,GUGAAU,GUUUUG,UGAAUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPA3 | 3 | 2140 | 0.015564202 | 0.003102746 | 2.326614 | AAGGAG,GCCAAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| EIF4B | 1 | 1179 | 0.007782101 | 0.001710061 | 2.186112 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPN1 | 1 | 1222 | 0.007782101 | 0.001772376 | 2.134475 | AGAAGA | AAAAGA,AGAAGA |
| PUM2 | 2 | 1890 | 0.011673152 | 0.002740445 | 2.090712 | GUACAU,UACAUC | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| YBX2 | 1 | 1480 | 0.007782101 | 0.002146271 | 1.858327 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NONO | 1 | 1498 | 0.007782101 | 0.002172357 | 1.840899 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| PABPC5 | 2 | 2400 | 0.011673152 | 0.003479539 | 1.746226 | AGAAAG,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| G3BP2 | 1 | 1644 | 0.007782101 | 0.002383941 | 1.706812 | GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| AGO2 | 1 | 1830 | 0.007782101 | 0.002653492 | 1.552267 | AAAGUG | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SRSF4 | 3 | 3740 | 0.015564202 | 0.005421472 | 1.521475 | AGAAGA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| CSTF2 | 1 | 1967 | 0.007782101 | 0.002852033 | 1.448169 | GUUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ZRANB2 | 2 | 3173 | 0.011673152 | 0.004599773 | 1.343559 | AAAGGU,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| HNRNPA1 | 16 | 18070 | 0.066147860 | 0.026188565 | 1.336757 | AAGAGG,AAGGAG,AAGGUG,AGAUUA,AGUGAA,CAGGGA,GAGGAA,GAGUGG,GCCAAG,GGAAGG,GGCAGG,GGGCAG,UAGGCA,UAGUGA,UUUAUA | AAAAAA,AAAGAG,AAGAGG,AAGGAG,AAGGUG,AAGUAC,AAUUUA,ACUAGA,AGACUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUACG,AGUAGG,AGUGAA,AGUGGU,AGUUAG,AUAGAA,AUAGCA,AUAGGG,AUUAGA,AUUUAA,AUUUAU,CAAAGA,CAAGGA,CAGGGA,CCAAGG,CCCCCC,CGUAGG,CUAGAC,GAAGGU,GACUAG,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGCC,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGGUUA,GGUGCG,GGUUAG,GUAAGU,GUACGC,GUAGGG,GUAGGU,GUGGUG,GUUAGG,GUUUAG,UAAGUA,UACGGC,UAGACA,UAGACU,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGAU,UAGGCA,UAGGCU,UAGGGA,UAGGGC,UAGGGU,UAGGUA,UAGGUC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUAUUU,UUUAGA,UUUAUA,UUUAUU,UUUUUU |
| RBM3 | 1 | 2152 | 0.007782101 | 0.003120136 | 1.318551 | GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| TRA2A | 5 | 6871 | 0.023346304 | 0.009958930 | 1.229132 | AAGAGG,AGAAAG,AGAAGA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PUM1 | 4 | 5823 | 0.019455253 | 0.008440164 | 1.204817 | ACAUAA,CCAGAA,GUACAU,UACAUC | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RBM24 | 1 | 2357 | 0.007782101 | 0.003417223 | 1.187335 | GAGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| FUS | 2 | 3648 | 0.011673152 | 0.005288145 | 1.142360 | CGGUGG,UGGUGG | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| SRSF5 | 6 | 8869 | 0.027237354 | 0.012854439 | 1.083320 | AGAAGA,CAACAG,GAGGAA,GGAAGA,UACAUC,UGCAGA | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| SART3 | 1 | 2634 | 0.007782101 | 0.003818652 | 1.027096 | GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
RBP on BSJ (Exon Only)
Plot
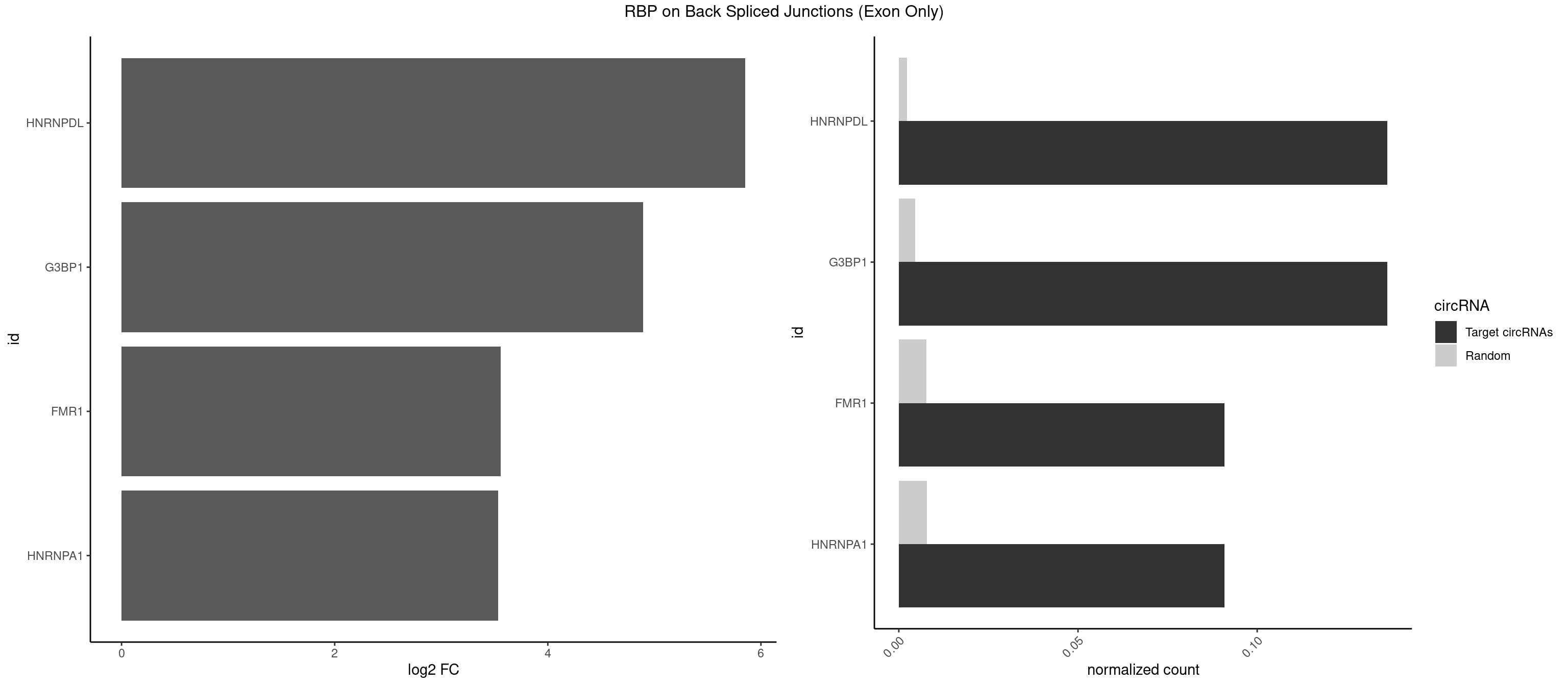
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPDL | 2 | 35 | 0.13636364 | 0.002357873 | 5.853829 | CUAGGC,UAGGCA | AACAGC,AAUACC,AAUUUA,ACACCA,ACAGCA,ACCAGA,AGAUAU,AGUAGG,AUCUGA,AUUAGC,AUUAGG,CACGCA,CCAGAC,CUAAGC,CUAAGU,CUAGAU,CUAGGA,CUAGGC,CUUUAG,GAACUA,GAUUAG,GCACUA,GCUAGU,UGCGCA,UUAGCC,UUAGGC,UUUAGG |
| G3BP1 | 2 | 69 | 0.13636364 | 0.004584752 | 4.894471 | CUAGGC,UAGGCA | ACAGGC,ACCCCC,ACCCCU,ACGCAG,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUCCGC,CACAGG,CACCCG,CACCGG,CACGCA,CAGGCA,CAGGCC,CAUCCG,CCACCC,CCAGGC,CCAUAC,CCAUCG,CCCACC,CCCAGG,CCCAUC,CCCCAC,CCCCGC,CCCCUC,CCCUCC,CCCUCG,CCUAGG,CCUCCG,CUACGC,CUAGGC,UACGCA,UCCGCC |
| FMR1 | 1 | 117 | 0.09090909 | 0.007728583 | 3.556149 | UAGGCA | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
| HNRNPA1 | 1 | 119 | 0.09090909 | 0.007859576 | 3.531901 | UAGGCA | AAAAAA,AAAGAG,AAGAGG,AAGGUG,AAUUUA,AGAGGA,AGAUAU,AGAUUA,AGGAAG,AGGAGC,AGGGAC,AGGGCA,AGGGUU,AGUAGG,AGUGAA,AUAGGG,AUUAGA,AUUUAA,CAAAGA,CAAGGA,CAGGGA,CCAAGG,GAAGGU,GACUUA,GAGGAA,GAGGAG,GAGUGG,GAUUAG,GCAGGC,GCCAAG,GGAAGG,GGACUU,GGAGGA,GGCAGG,GGGACU,GGGCAG,GGUGCG,GUUAGG,UAGACA,UAGAGA,UAGAGU,UAGAUU,UAGGAA,UAGGCU,UAGGGC,UAGUGA,UAGUUA,UCGGGC,UGGUGC,UUAGAU,UUAGGG,UUUAGA |
RBP on BSJ (Exon and Intron)
Plot
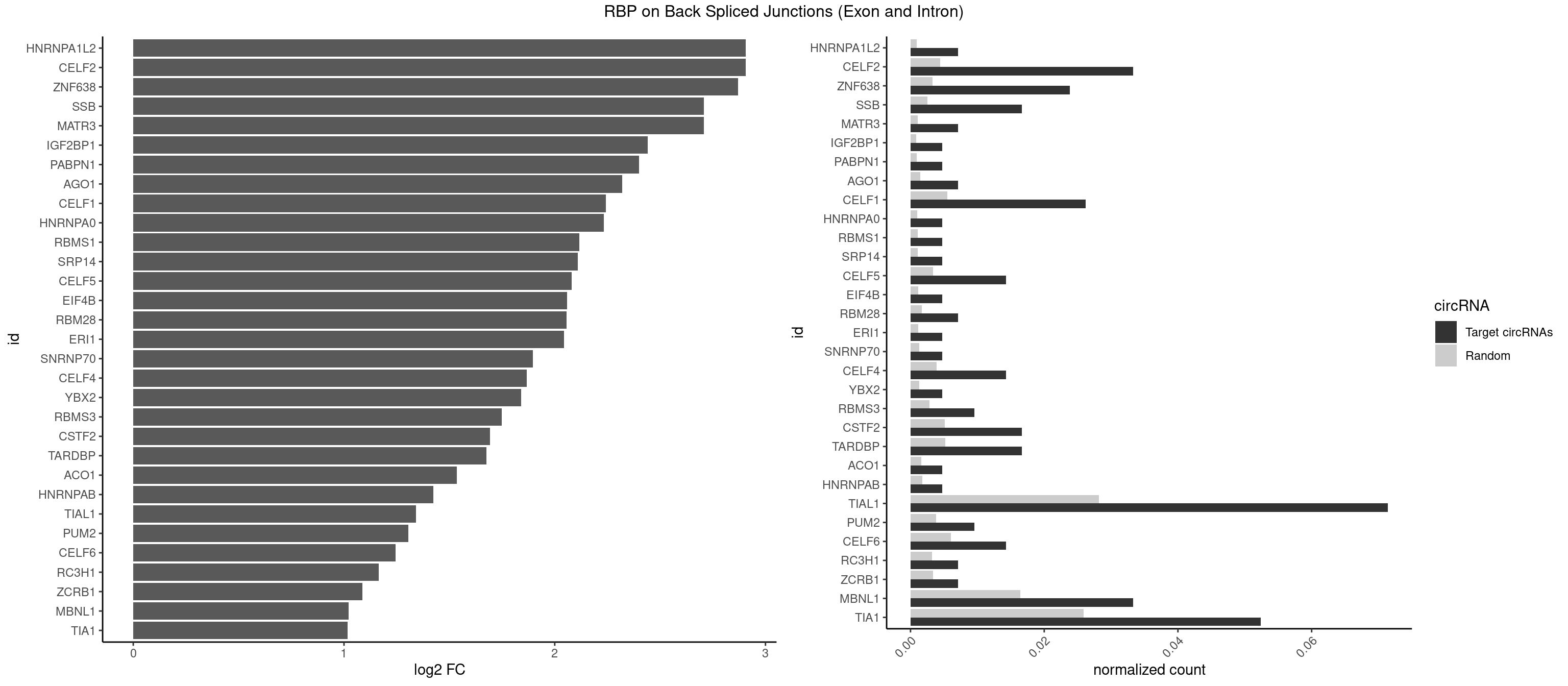
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | GUAGGG,UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| CELF2 | 13 | 881 | 0.033333333 | 0.0044437727 | 2.907109 | AUGUGU,GUAUGU,GUUGUU,UAUGUG,UGUGUG,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ZNF638 | 9 | 646 | 0.023809524 | 0.0032597743 | 2.868695 | GUUGUU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SSB | 6 | 505 | 0.016666667 | 0.0025493753 | 2.708750 | CUGUUU,GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | AGGUAG,GGUAGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| CELF1 | 10 | 1097 | 0.026190476 | 0.0055320435 | 2.243158 | CUGUCU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | AUAUAG | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| CELF5 | 5 | 669 | 0.014285714 | 0.0033756550 | 2.081334 | GUGUGG,GUGUUU,UGUGUG,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF4 | 5 | 776 | 0.014285714 | 0.0039147521 | 1.867580 | GUGUGG,GUGUUU,UGUGUG,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAG,CAUAUA,UAUAGC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| CSTF2 | 6 | 1022 | 0.016666667 | 0.0051541717 | 1.693153 | GUGUUU,UGUGUG,UGUGUU,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 6 | 1034 | 0.016666667 | 0.0052146312 | 1.676328 | GUUGUU,UGUGUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| TIAL1 | 29 | 5597 | 0.071428571 | 0.0282043531 | 1.340583 | AAAUUU,AAUUUU,AGUUUU,AUUUUC,AUUUUG,AUUUUU,CUUUUG,GUUUUU,UAAAUU,UAUUUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUCAG,UUUUAA,UUUUCA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | GUACAU,UACAUC,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| CELF6 | 5 | 1196 | 0.014285714 | 0.0060308343 | 1.244144 | GUGUGG,UGUGGG,UGUGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| MBNL1 | 13 | 3258 | 0.033333333 | 0.0164197904 | 1.021530 | AUGCUU,CUGCCA,CUGCCU,CUGCUG,CUUGCU,GCUGCU,GCUUGC,GUGCUU,UGCUGU,UGCUUU,UUGCCC,UUGCUG | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| TIA1 | 21 | 5140 | 0.052380952 | 0.0259018541 | 1.015987 | AUUUUC,AUUUUG,AUUUUU,CUUUUG,GUUUUA,GUUUUU,UAUUUU,UUAUUU,UUUUCU,UUUUGU,UUUUUA,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.