circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000185842:+:1:225264196:225290082
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000185842:+:1:225264196:225290082 | ENSG00000185842 | ENST00000682510 | + | 1 | 225264197 | 225290082 | 1312 | UAGCCUAAAACAGAAUAUCACCAUCUUGAUUCCUGAAACUCAUAAGACAGCAACUGGAAGUUCAGAUAAUCCCACUAAAAAGCCAGAAGUUAGAACUAAUAAAAAGUUACUUAAAAAUAAUGAUCAUAAAGGAGUUGUAGUCUCUACAAUAAAUUUUAGCACCAAUGUAACAGCUGCCAAAACCAAGGAGAUGAUUCUUAAGAAGUUAAUAAGAAGAACUAAAGAUACUCUUGGAGCACCAAAAAACAACCGGAUUCUAAUAUUUAUUGAUGAUAUGAAUAUGCCAGUAUCAGAUAUGUAUGGAGCACAGCCACCCCUGGAAUUGAUAAGACAAUUGUUAGAUUUGGGAGGAGUUUAUGAUACUGAAAAAAAUACAUGGAAGAAUAUUCAAGAUCUGUCUAUAGUUGCAGCUUGUGUUCCAGUUGUGAAUGAUAUCAGCCCACGUCUUCUCAAACACUUUUCCAUGCUGGUAUUACCUCAUCCUUCACAAGACAUCUUAUGUACUAUUUUCCAGGCUCAUUUGGGAAUUUAUUUCUCCAUCAAUAACUUCACACCUGAAGUUCAGAAAAGUAAGGAUCAGAUAAUAUCUUGUUCCCUAGCUAUAUACCAUCAAGUACGUCAGAAUAUGUUACCAACUCCAACAAAAUGUCACUACAUGUUUAAUCUUCGAGAUAUGUUUAAGCUUCUCCUAGGAUUGCUGCAAGCUGACAGGACUGUUGUUAACUCCAAAGAGAUGGCUGCUCUGCUCUUUGUUCAUGAAGCCACCCGAGUAUUUCACGAUCGCUUAAUUGAUUUCACUGAUAAAAGCCUUUUCUAUCGGUUGCUUUCAAGGGAACUUGAGAACUGUUUUCAGAUUCAGUGGACCCAAGAAAACCUGAUGAAUCACUCAACUGUAUUUUUGGACUUCUUGGAUAUAAACAAGACUCAUAGAAAAAAGAUUUAUCAGAAUACCAGUGACUAUAAUAAACUUGCCAGUGUACUUGAUGAAUUCCAAAUGAAGUUGGGUUCAAUUUCUUUGGAGCUUUCUCAUUCCAUGGUGUUCUUCAAGGAAGCCAUAGAACACAUCAUAAGAGCAACAAGGGUUCUUCGACAACCAGGGAGUCACAUGUUACUGAUUGGAAUAGAUGGAUGUGGGAAAAAAACAUGUGCAACCUUGGCCUGUUAUUUGACAGAUAAUAAACUAUACCGAGUGCCUAUAUCUCACAAAUGUGCCUACAUCGAAUUCAAAGAAGUCUUUAAAAAGGUGUUUAUUCACGCAGGAUUAAAAGGGAAACCCACUGUUCUGAUGGUUCCCAAUUUAAACAUAGAACAAUAGCCUAAAACAGAAUAUCACCAUCUUGAUUCCUGAAACUCAUAAGACAG | circ |
| ENSG00000185842:+:1:225264196:225290082 | ENSG00000185842 | ENST00000682510 | + | 1 | 225264197 | 225290082 | 22 | ACAUAGAACAAUAGCCUAAAAC | bsj |
| ENSG00000185842:+:1:225264196:225290082 | ENSG00000185842 | ENST00000682510 | + | 1 | 225263997 | 225264206 | 210 | GUUAGCUUAUGUUUUGUUCAGUUUGGUAUAAGUUCUUAUGGUGACACUAAUUUCAUCUGAAAAGGUGGAAACAGAAAAAAAAUUUUUCUUGUAAGAGUUAGAAGUAAAAUUCACAAUAUGCUGUUACUUUUUUGUUGUUGUUUAAUAUACCUAUUAUGGUAAACAAACUAAUUUUGAAUCCUUAAAAUAUUUCAUUCCAGUAGCCUAAAA | ie_up |
| ENSG00000185842:+:1:225264196:225290082 | ENSG00000185842 | ENST00000682510 | + | 1 | 225290073 | 225290282 | 210 | CAUAGAACAAGUAAGUACUUUUUGUCUUUGCUAUUUGCUGAAGUUUGAUAUCUAUGUGGCUACCCCCUCCCCAAAAAUUUAAUAUUUGAAAAGAAAACAAGAAAAUAUUUAUCAAUGGUUCUAUUACCCUACAAUAUCAGUUUUCUUGCAGUAUUCUCUGUUCUUAUCUAGUCAUAGCCCACACAGUUUAUAUAUUUGUAAUGACGCCAU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
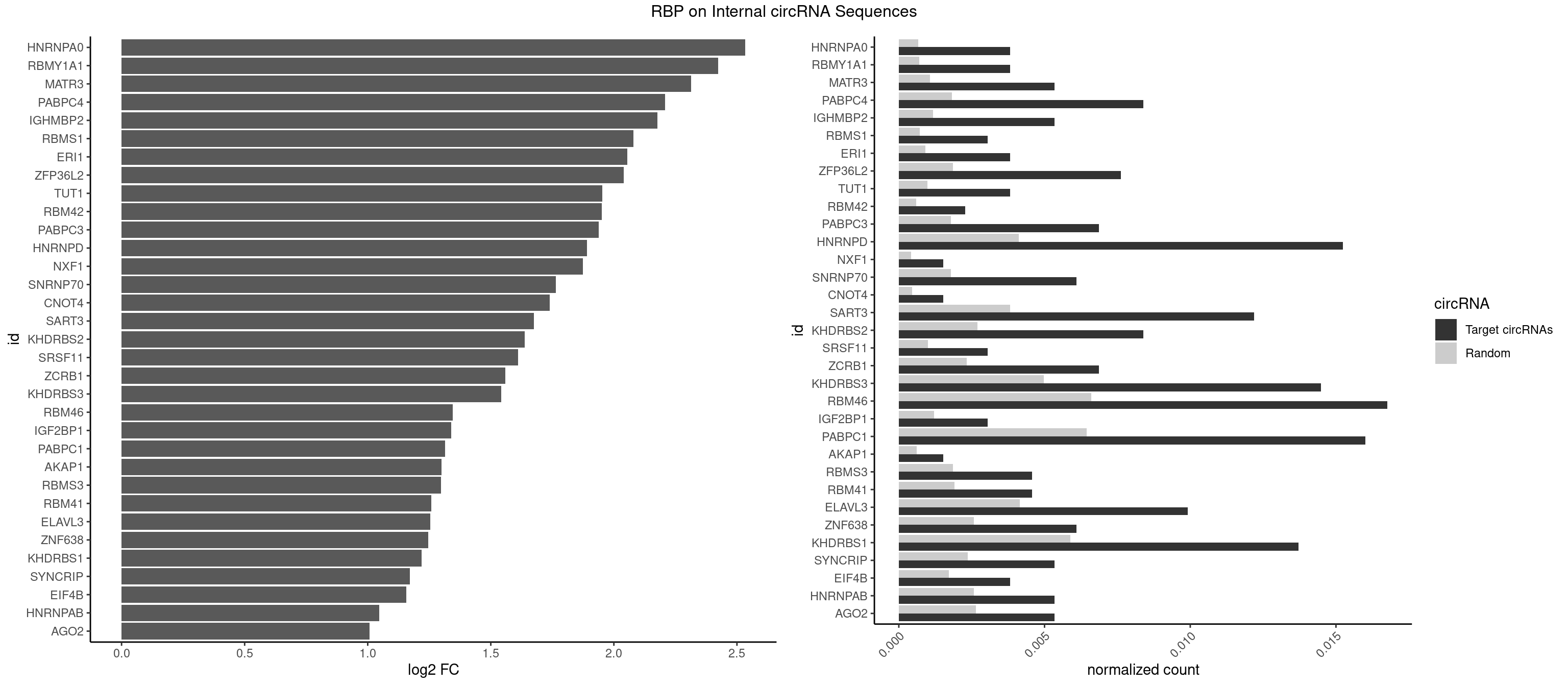
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA0 | 4 | 453 | 0.003810976 | 0.0006579386 | 2.534136 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 4 | 489 | 0.003810976 | 0.0007101099 | 2.424046 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| MATR3 | 6 | 739 | 0.005335366 | 0.0010724109 | 2.314729 | AAUCUU,AUCUUA,AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PABPC4 | 10 | 1251 | 0.008384146 | 0.0018144033 | 2.208169 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| IGHMBP2 | 6 | 813 | 0.005335366 | 0.0011796520 | 2.177226 | AAAAAA | AAAAAA |
| RBMS1 | 3 | 497 | 0.003048780 | 0.0007217036 | 2.078754 | AUAUAC,GAUAUA,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ERI1 | 4 | 632 | 0.003810976 | 0.0009173461 | 2.054622 | UUCAGA,UUUCAG | UUCAGA,UUUCAG |
| ZFP36L2 | 9 | 1277 | 0.007621951 | 0.0018520827 | 2.041012 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| TUT1 | 4 | 678 | 0.003810976 | 0.0009840095 | 1.953416 | AAAUAC,AGAUAC,GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM42 | 2 | 407 | 0.002286585 | 0.0005912752 | 1.951293 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| PABPC3 | 8 | 1234 | 0.006859756 | 0.0017897669 | 1.938386 | AAAAAC,AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| HNRNPD | 19 | 2837 | 0.015243902 | 0.0041128408 | 1.890025 | AAAAAA,AAUUUA,AGAUAU,AUUUAU,UAUUUA,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| NXF1 | 1 | 286 | 0.001524390 | 0.0004159215 | 1.873849 | AACCUG | AACCUG |
| SNRNP70 | 7 | 1237 | 0.006097561 | 0.0017941145 | 1.764960 | AUCAAG,AUUCAA,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CNOT4 | 1 | 314 | 0.001524390 | 0.0004564992 | 1.739548 | GACAGA | GACAGA |
| SART3 | 15 | 2634 | 0.012195122 | 0.0038186524 | 1.675169 | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| KHDRBS2 | 10 | 1858 | 0.008384146 | 0.0026940701 | 1.637876 | AAUAAA,AUAAAA,AUAAAC,GAUAAA | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| SRSF11 | 3 | 688 | 0.003048780 | 0.0009985015 | 1.610396 | AAGAAG | AAGAAG |
| ZCRB1 | 8 | 1605 | 0.006859756 | 0.0023274215 | 1.559425 | ACUUAA,AUUUAA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| KHDRBS3 | 18 | 3429 | 0.014481707 | 0.0049707696 | 1.542691 | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UUAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| RBM46 | 21 | 4554 | 0.016768293 | 0.0066011240 | 1.344952 | AAUGAA,AAUGAU,AUCAAG,AUCAAU,AUCAUA,AUGAAG,AUGAAU,AUGAUA,AUGAUU,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| IGF2BP1 | 3 | 831 | 0.003048780 | 0.0012057377 | 1.338316 | AGCACC,CACCCG | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PABPC1 | 20 | 4443 | 0.016006098 | 0.0064402624 | 1.313430 | AAAAAA,AAAAAC,ACAAAU,ACUAAU,AGAAAA,CAAACA,CUAAUA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| AKAP1 | 1 | 426 | 0.001524390 | 0.0006188101 | 1.300664 | UAUAUA | AUAUAU,UAUAUA |
| RBMS3 | 5 | 1283 | 0.004573171 | 0.0018607779 | 1.297289 | CUAUAG,CUAUAU,UAUAUA,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM41 | 5 | 1318 | 0.004573171 | 0.0019115000 | 1.258490 | AUACAU,UACAUG,UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| ELAVL3 | 12 | 2867 | 0.009908537 | 0.0041563169 | 1.253366 | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| ZNF638 | 7 | 1773 | 0.006097561 | 0.0025708878 | 1.245966 | GGUUCU,GUUCUU,GUUGUU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| KHDRBS1 | 17 | 4064 | 0.013719512 | 0.0058910141 | 1.219641 | AUAAAA,AUUUAA,CAAAAU,CUAAAA,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| SYNCRIP | 6 | 1634 | 0.005335366 | 0.0023694485 | 1.171036 | AAAAAA | AAAAAA,UUUUUU |
| EIF4B | 4 | 1179 | 0.003810976 | 0.0017100607 | 1.156113 | CUUGGA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPAB | 6 | 1782 | 0.005335366 | 0.0025839306 | 1.046020 | AAGACA,AGACAA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| AGO2 | 6 | 1830 | 0.005335366 | 0.0026534924 | 1.007695 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
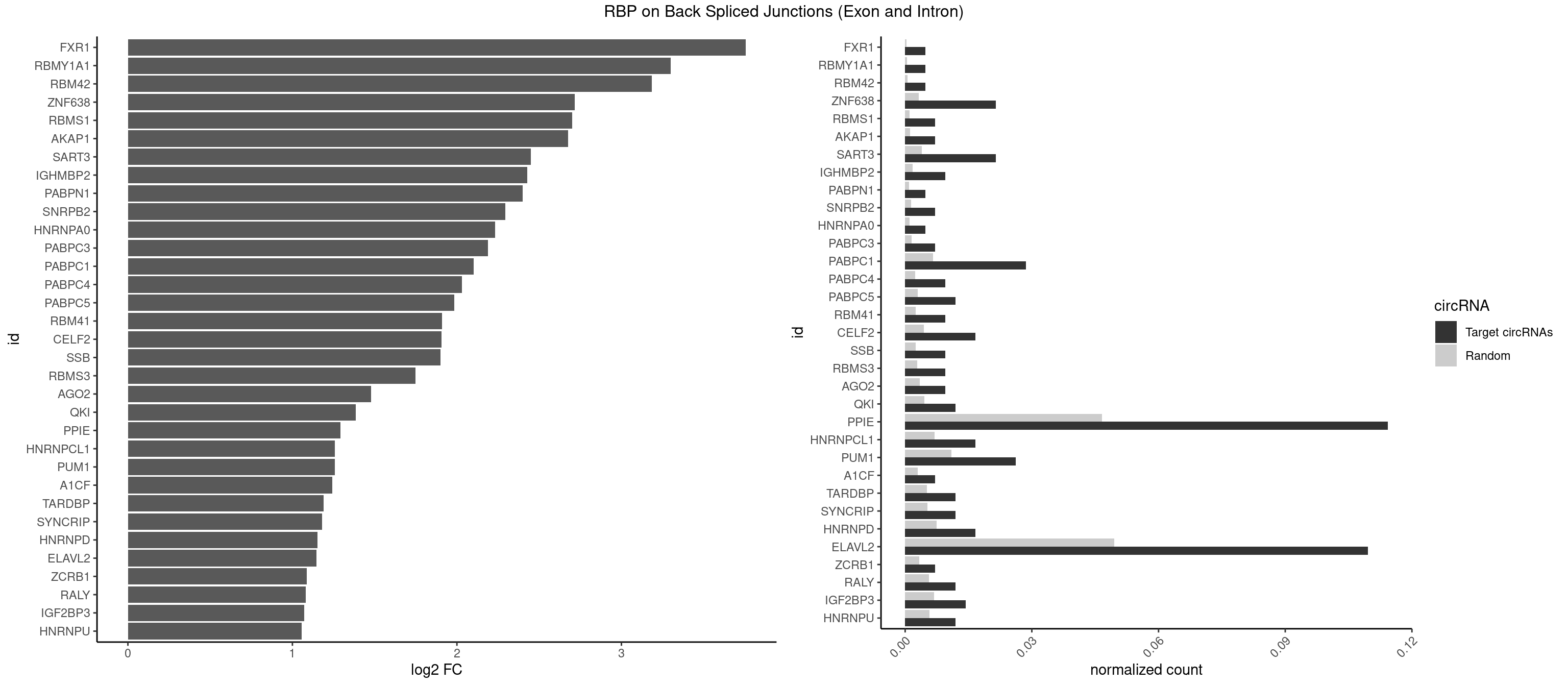
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | ACAAGA | ACAAGA,CAAGAC |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| ZNF638 | 8 | 646 | 0.021428571 | 0.0032597743 | 2.716692 | GGUUCU,GUUCUU,GUUGUU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | AUAUAC,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 2 | 221 | 0.007142857 | 0.0011185006 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| SART3 | 8 | 777 | 0.021428571 | 0.0039197904 | 2.450687 | AAAAAA,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| IGHMBP2 | 3 | 350 | 0.009523810 | 0.0017684401 | 2.429061 | AAAAAA | AAAAAA |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC1 | 11 | 1321 | 0.028571429 | 0.0066606207 | 2.100845 | AAAAAA,ACAAAC,ACUAAU,AGAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| PABPC4 | 3 | 462 | 0.009523810 | 0.0023327287 | 2.029520 | AAAAAA | AAAAAA,AAAAAG |
| PABPC5 | 4 | 596 | 0.011904762 | 0.0030078597 | 1.984730 | AGAAAA,GAAAAU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| CELF2 | 6 | 881 | 0.016666667 | 0.0044437727 | 1.907109 | GUUGUU,UAUGUG,UAUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SSB | 3 | 505 | 0.009523810 | 0.0025493753 | 1.901395 | GCUGUU,UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AAUAUA,AUAUAU,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| AGO2 | 3 | 677 | 0.009523810 | 0.0034159613 | 1.479247 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | ACACUA,ACUAAU,CACUAA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| PPIE | 47 | 9262 | 0.114285714 | 0.0466696896 | 1.292087 | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAU,AAAUUU,AAUAUA,AAUAUU,AAUUUA,AAUUUU,AUAUAU,AUAUUU,AUUUAA,AUUUAU,AUUUUU,UAAAAU,UAAUAU,UAAUUU,UAUAUA,UAUAUU,UAUUAU,UAUUUA,UUAAAA,UUAAUA,UUAUAU,UUUAAU,UUUAUA,UUUUUU | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| HNRNPCL1 | 6 | 1381 | 0.016666667 | 0.0069629182 | 1.259202 | AUUUUU,CUUUUU,UUUUUG,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| PUM1 | 10 | 2172 | 0.026190476 | 0.0109482064 | 1.258348 | AAUAUU,CUUGUA,UAAUAU,UAUAUA,UGUAAU,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| A1CF | 2 | 598 | 0.007142857 | 0.0030179363 | 1.242939 | AUCAGU,CAGUAU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUUGUU,GUUUUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SYNCRIP | 4 | 1041 | 0.011904762 | 0.0052498992 | 1.181177 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.0075020153 | 1.151615 | AAAAAA,AAUUUA,AUUUAU,UAUUUA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| ELAVL2 | 45 | 9826 | 0.109523810 | 0.0495112858 | 1.145415 | AAUUUA,AAUUUU,AUAUUU,AUUUAA,AUUUAU,AUUUUG,AUUUUU,CUUUUU,GUUUUC,UAAGUU,UAAUUU,UACUUU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUUA,UAUUUG,UCUUAU,UUACUU,UUAUAU,UUAUGU,UUCAUU,UUUAAU,UUUAUA,UUUCAU,UUUCUU,UUUGAU,UUUUUC,UUUUUG,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AUUUAA,GUUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUG,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAACA,AAAUUC,AAUUCA,ACAAAC,CACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| HNRNPU | 4 | 1136 | 0.011904762 | 0.0057285369 | 1.055300 | AAAAAA,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.