circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000081760:+:12:125102678:125107268
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000081760:+:12:125102678:125107268 | ENSG00000081760 | ENST00000316519 | + | 12 | 125102679 | 125107268 | 345 | GGUGUGCUGGACCGGUUUUCUCAAAUUCAGCCAAAGCUCAUCUUCUCUGUGGAGGCUGUUGUCUAUAAUGGCAAAGAGCACAACCACAUGGAAAAGCUGCAGCAGGUGGUUAAAGGCCUACCAGACUUGAAGAAAGUGGUGGUGAUUCCUUAUGUGUCCUCCAGAGAGAACAUAGACCUUUCAAAGAUUCCAAACAGUGUGUUUCUGGAUGACUUUCUUGCCACCGGCACCAGUGAGCAGGCCCCGCAGCUGGAGUUCGAGCAGCUGCCCUUCAGCCACCCACUGUUCAUCAUGUUCUCAUCGGGCACCACGGGCGCACCCAAGUGCAUGGUGCAUUCCGCUGGGGGUGUGCUGGACCGGUUUUCUCAAAUUCAGCCAAAGCUCAUCUUCUCUGU | circ |
| ENSG00000081760:+:12:125102678:125107268 | ENSG00000081760 | ENST00000316519 | + | 12 | 125102679 | 125107268 | 22 | AUUCCGCUGGGGGUGUGCUGGA | bsj |
| ENSG00000081760:+:12:125102678:125107268 | ENSG00000081760 | ENST00000316519 | + | 12 | 125102479 | 125102688 | 210 | UUUUUAGAGACAGGGUCUCACUCGUGCCCAGGCUGGAAUGCAGUGGUGCAAGCAUAGCUCACUCCAACCUUGAACUCCUGGGCUCAAGCAAUCCUCCCACCUCAGCCUCCAGAGUAGCUGGGACUACAGGCACCCACCACCAUGCCUGCUGGUUUUUGCCUUUUGGAUGAUCAAUGAUGACUUUUUCCUCCUGCUUUCAGGGUGUGCUGG | ie_up |
| ENSG00000081760:+:12:125102678:125107268 | ENSG00000081760 | ENST00000316519 | + | 12 | 125107259 | 125107468 | 210 | UUCCGCUGGGGUAGGUCUCUGGGGAAGGCUCUGCAGGGCCUCCUGUUGUCUGUUUGCUUCAACAGGGCCUCCCAGCUGAUGAUGGCAGUGUUGGAGUUGUCAAACUGCACCCCAUCCCCGGAGAGUCCAACGUGCAGCUUCUCUCCAAGUGGGGUGCUGCACGGAGAUCCGGCACUGGGCCCAGGGCAGUGGAGUGCUGCCACUUCAAAG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
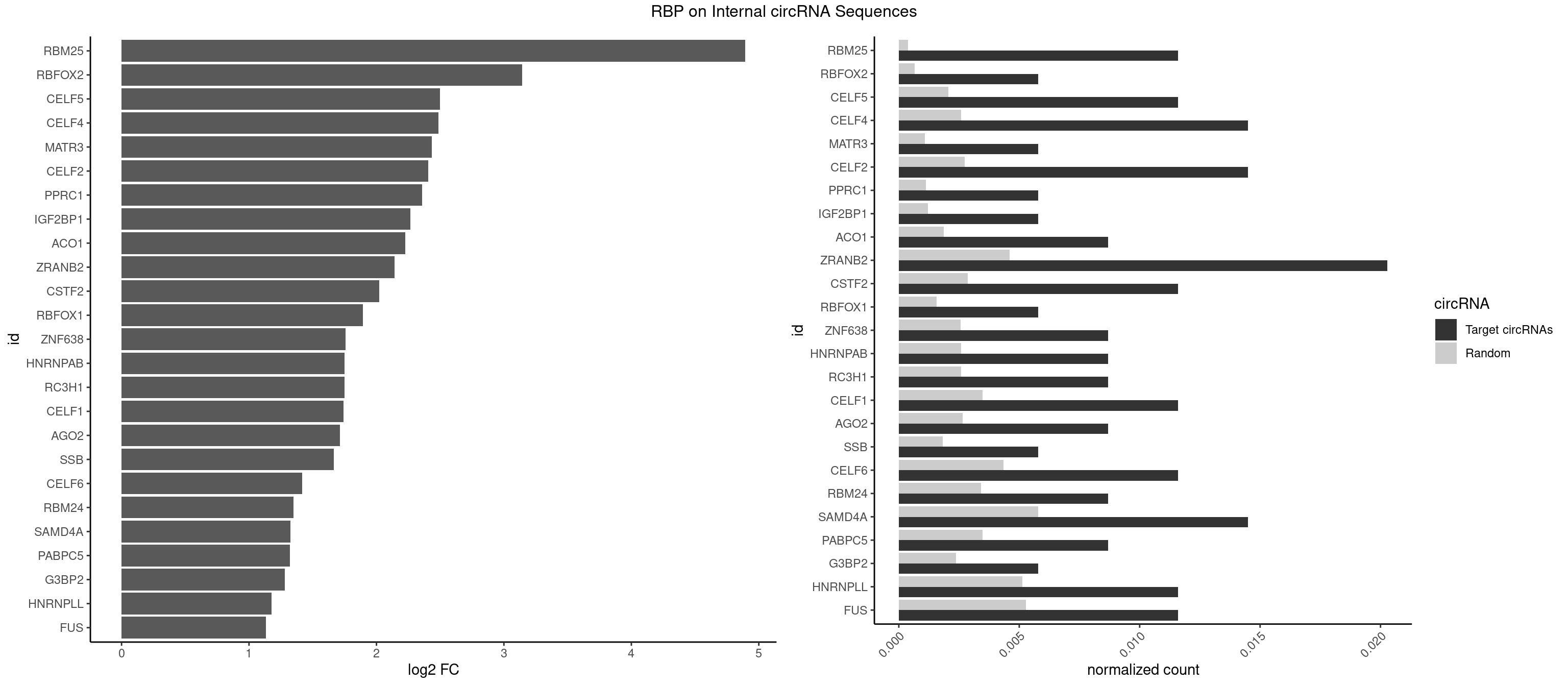
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 3 | 268 | 0.011594203 | 0.0003898359 | 4.894393 | AUCGGG,CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| RBFOX2 | 1 | 452 | 0.005797101 | 0.0006564894 | 3.142488 | UGCAUG | UGACUG,UGCAUG |
| CELF5 | 3 | 1415 | 0.011594203 | 0.0020520728 | 2.498250 | GUGUGU,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 4 | 1782 | 0.014492754 | 0.0025839306 | 2.487692 | GGUGUG,GUGUGU,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| MATR3 | 1 | 739 | 0.005797101 | 0.0010724109 | 2.434474 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CELF2 | 4 | 1886 | 0.014492754 | 0.0027346479 | 2.405905 | AUGUGU,GUGUGU,UAUGUG,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PPRC1 | 1 | 780 | 0.005797101 | 0.0011318283 | 2.356677 | GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| IGF2BP1 | 1 | 831 | 0.005797101 | 0.0012057377 | 2.265416 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ACO1 | 2 | 1283 | 0.008695652 | 0.0018607779 | 2.224388 | CAGUGA,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZRANB2 | 6 | 3173 | 0.020289855 | 0.0045997733 | 2.141124 | GGUGGU,GUGGUG,UAAAGG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| CSTF2 | 3 | 1967 | 0.011594203 | 0.0028520334 | 2.023341 | GUGUGU,GUGUUU,UGUGUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBFOX1 | 1 | 1077 | 0.005797101 | 0.0015622419 | 1.891714 | UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| ZNF638 | 2 | 1773 | 0.008695652 | 0.0025708878 | 1.758028 | UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPAB | 2 | 1782 | 0.008695652 | 0.0025839306 | 1.750727 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RC3H1 | 2 | 1786 | 0.008695652 | 0.0025897275 | 1.747494 | CCCUUC,UCUGUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CELF1 | 3 | 2391 | 0.011594203 | 0.0034664959 | 1.741854 | GUGUGU,UGUGUU,UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| AGO2 | 2 | 1830 | 0.008695652 | 0.0026534924 | 1.712402 | AAAGUG,AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SSB | 1 | 1260 | 0.005797101 | 0.0018274462 | 1.665503 | GCUGUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| CELF6 | 3 | 2997 | 0.011594203 | 0.0043447134 | 1.416071 | GUGGUG,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM24 | 2 | 2357 | 0.008695652 | 0.0034172229 | 1.347470 | AGUGUG,GUGUGU | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SAMD4A | 4 | 3992 | 0.014492754 | 0.0057866714 | 1.324526 | CGGGCA,CUGGAC,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC5 | 2 | 2400 | 0.008695652 | 0.0034795387 | 1.321398 | AGAAAG,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| G3BP2 | 1 | 1644 | 0.005797101 | 0.0023839405 | 1.281984 | GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPLL | 3 | 3534 | 0.011594203 | 0.0051229360 | 1.178361 | ACCACA,CAAACA,CACCAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| FUS | 3 | 3648 | 0.011594203 | 0.0052881452 | 1.132570 | GGGUGU,UGGUGA,UGGUGG | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
RBP on BSJ (Exon Only)
Plot
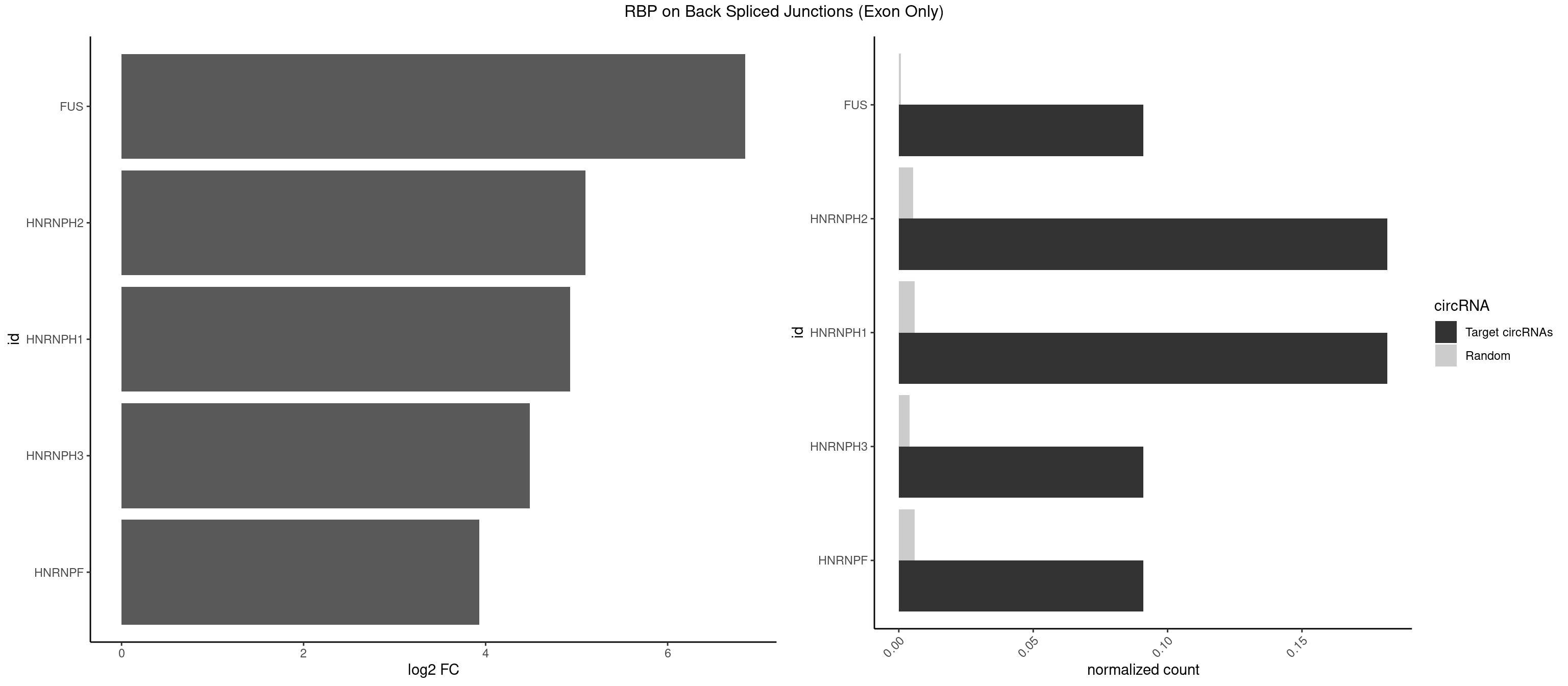
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FUS | 1 | 11 | 0.09090909 | 0.0007859576 | 6.853829 | GGGUGU | AAAAAA,CGGUGG,GGGUGA,GGGUGG,GGGUGU |
| HNRNPH2 | 3 | 80 | 0.18181818 | 0.0053052135 | 5.098942 | CUGGGG,GGGUGU,UGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPH1 | 3 | 90 | 0.18181818 | 0.0059601782 | 4.930997 | CUGGGG,GGGUGU,UGGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPH3 | 1 | 61 | 0.09090909 | 0.0040607807 | 4.484596 | GGGUGU | AAGGGA,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CGAGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGUG,UGUGGG |
| HNRNPF | 1 | 90 | 0.09090909 | 0.0059601782 | 3.930997 | CUGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
RBP on BSJ (Exon and Intron)
Plot
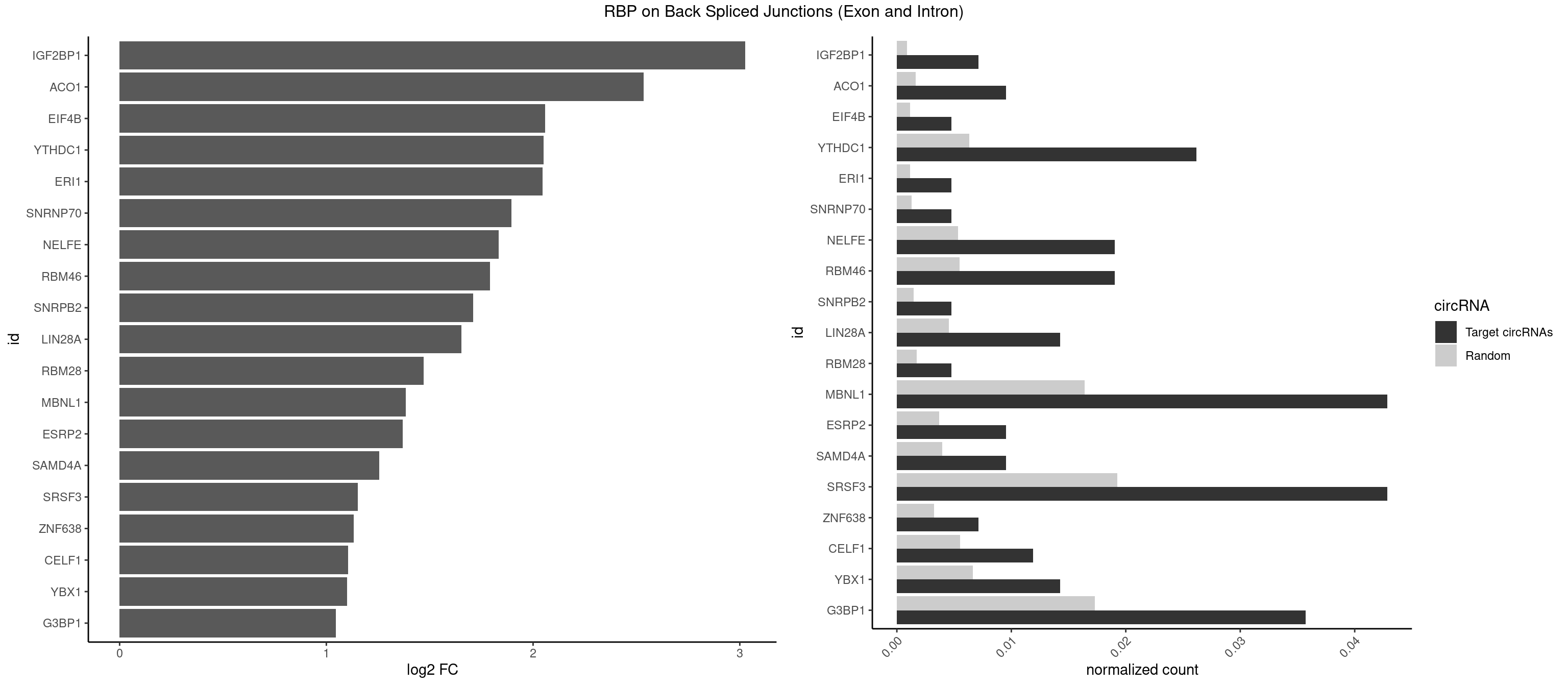
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| IGF2BP1 | 2 | 173 | 0.007142857 | 0.0008766626 | 3.026408 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ACO1 | 3 | 325 | 0.009523810 | 0.0016424829 | 2.535660 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| YTHDC1 | 10 | 1254 | 0.026190476 | 0.0063230552 | 2.050349 | GAAUGC,GACUAC,GAGUGC,GCCUGC,GGGUGC,UCCUGC,UCGUGC,UGCUGC,UGGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| NELFE | 7 | 1059 | 0.019047619 | 0.0053405885 | 1.834540 | CUCUGG,CUGGUU,GGUCUC,GUCUCU,UCUCUC,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM46 | 7 | 1091 | 0.019047619 | 0.0055018138 | 1.791631 | AAUGAU,AUCAAU,AUGAUG,GAUCAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| LIN28A | 5 | 900 | 0.014285714 | 0.0045395002 | 1.653968 | CGGAGA,GGAGAU,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| MBNL1 | 17 | 3258 | 0.042857143 | 0.0164197904 | 1.384100 | CCGCUG,CCUGCU,CUGCCA,CUGCUG,CUGCUU,GUGCUG,UCCGCU,UGCUGC,UGCUUC,UGCUUU,UUGCCU,UUGCUU,UUUGCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAG,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| SAMD4A | 3 | 790 | 0.009523810 | 0.0039852882 | 1.256855 | CUGGAA,GCUGGA,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| SRSF3 | 17 | 3825 | 0.042857143 | 0.0192765014 | 1.152692 | ACCACC,ACUACA,ACUUCA,CACCAC,CACUUC,CCACCA,CUACAG,CUCCAA,CUUCAA,GAUCAA,GCUUCA,UCUCCA,UGUCAA,UUCAAC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | UGUUGG,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF1 | 4 | 1097 | 0.011904762 | 0.0055320435 | 1.105654 | GUGUUG,UGUCUG,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | ACCACC,CACCAC,CCACCA,GCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| G3BP1 | 14 | 3431 | 0.035714286 | 0.0172914148 | 1.046445 | ACAGGC,ACCCAC,AGGCAC,CAGGCA,CCAGGC,CCAUCC,CCCACC,CCCAGG,CCCAUC,CCCCGG,CCGGCA,CGGCAC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.