circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000128683:+:2:170836792:170854022
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000128683:+:2:170836792:170854022 | ENSG00000128683 | ENST00000493875 | + | 2 | 170836793 | 170854022 | 978 | GUCAUCCUCGAUUUUUCAACCAGCUCUCCACUGGAUUGGAUAUUAUUGGCCUAGCUGGAGAAUGGCUGACAUCAACGGCCAAUACCAACAUGUUUACAUAUGAAAUUGCACCAGUGUUUGUCCUCAUGGAACAAAUAACACUUAAGAAGAUGAGAGAGAUAGUUGGAUGGUCAAGUAAAGAUGGUGAUGGGAUAUUUUCUCCUGGGGGCGCCAUAUCCAACAUGUACAGCAUCAUGGCUGCUCGCUACAAGUACUUCCCGGAAGUUAAGACAAAGGGCAUGGCGGCUGUGCCUAAACUGGUCCUCUUCACCUCAGAACAGAGUCACUAUUCCAUAAAGAAAGCUGGGGCUGCACUUGGCUUUGGAACUGACAAUGUGAUUUUGAUAAAGUGCAAUGAAAGGGGGAAAAUAAUUCCAGCUGAUUUUGAGGCAAAAAUUCUUGAAGCCAAACAGAAGGGAUAUGUUCCCUUUUAUGUCAAUGCAACUGCUGGCACGACUGUUUAUGGAGCUUUUGAUCCGAUACAAGAGAUUGCAGAUAUAUGUGAGAAAUAUAACCUUUGGUUGCAUGUCGAUGGAUUUAACUUCUCACAAUUGGCCAAUAGGAUCAUCUGCCUUGCUACUGAACUAAUGACUAACAAAGGCUGUGUCACGUGGCAUCCCAACUAUUCAGUAAACAUGCAUCAUGGCUGCCUGGGGAGGUGGGCUGCUCAUGUCCAGGAAGCACCGCCAUAAACUCAACGGCAUAGAAAGGGCCAACUCAGUCACCUGGAACCCUCACAAGAUGAUGGGCGUGCUGUUGCAGUGCUCUGCCAUUCUCGUCAAGGAAAAGGGUAUACUCCAAGGAUGCAACCAGAUGUGUGCAGGAUACCUCUUCCAGCCAGACAAGCAGUAUGAUGUCUCCUACGACACCGGGGACAAGGCAAUUCAGUGUGGCCGCCACGUGGAUAUCUUCAAGUUCUGGCUGAUGUGGAAAGCAAAGGUCAUCCUCGAUUUUUCAACCAGCUCUCCACUGGAUUGGAUAUUAUUGGC | circ |
| ENSG00000128683:+:2:170836792:170854022 | ENSG00000128683 | ENST00000493875 | + | 2 | 170836793 | 170854022 | 22 | GGAAAGCAAAGGUCAUCCUCGA | bsj |
| ENSG00000128683:+:2:170836792:170854022 | ENSG00000128683 | ENST00000493875 | + | 2 | 170836593 | 170836802 | 210 | UUUGAGGGACAUUAUGAAUGUUAAUGUUGGUAUAGGGUGGGUCAACCUGCCCUUCCUUUUUCAAUAUCCUUGAGCAGCCUUAUUCGACAUAUCACUGGGAUGGAAGCCCCAUCAGUGUGACCCAGUGGGGCAGGCCGUUUGCCUUCAAGAUAGGUUGAGAAAUCUGCAUUUGCCUUGAUGCUUUUCUGUUUCCUAUCUAGGUCAUCCUCG | ie_up |
| ENSG00000128683:+:2:170836792:170854022 | ENSG00000128683 | ENST00000493875 | + | 2 | 170854013 | 170854222 | 210 | GAAAGCAAAGGUAUGAAGGGAAUAGUUCAGGAAAUAGAGCAGAAAUGUAAUUUGCACAGACUGGCUGGCAAAAGAGGGGCAAAGAUAUAUCACAGAUAAUUCACUAUAUGGGGAAUCAAUAUCUAGAUAAUUGAAUUAACUGUGUUUUCUUUGUGAAUGAAAUUACUUUAUUCCUUAUGUCAAAUUCUCUCUCCUUUUGUUCAGUCUCCU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
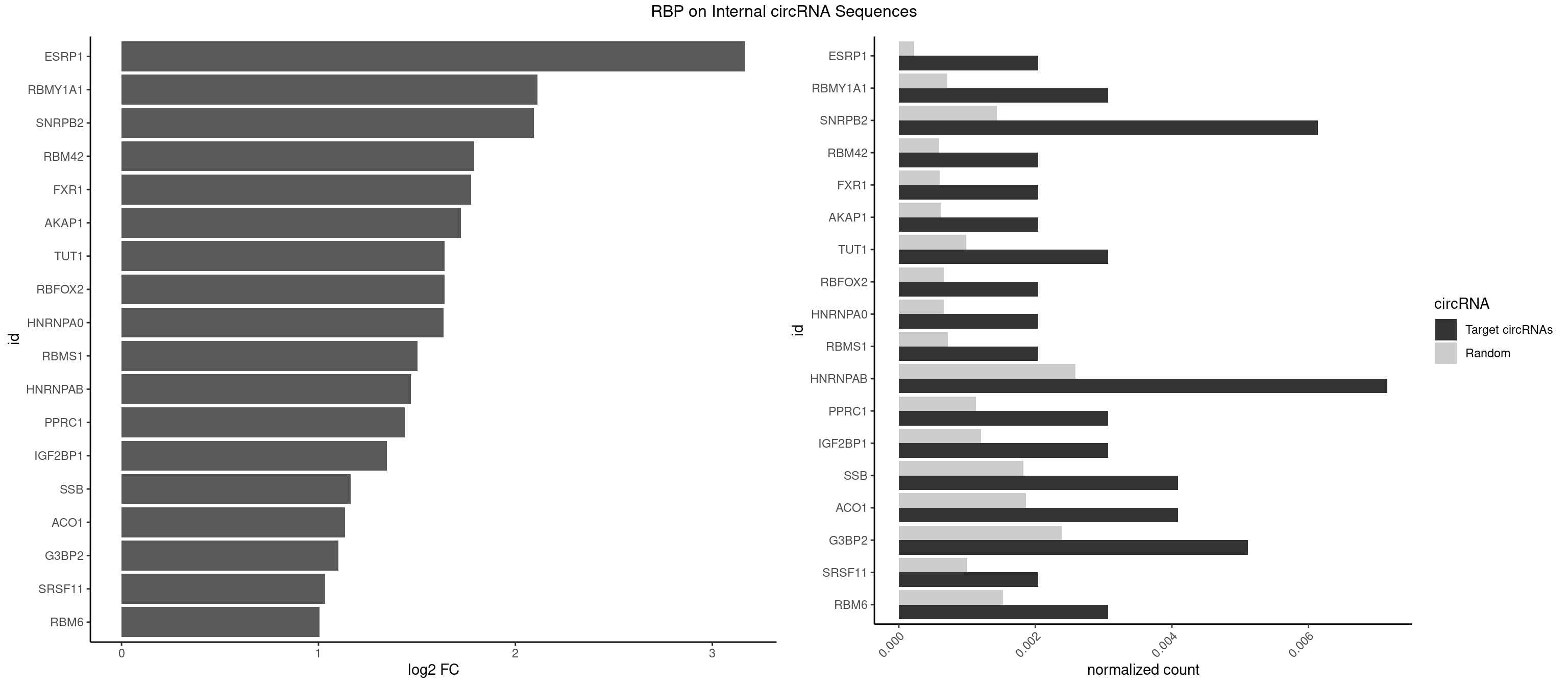
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.002044990 | 0.0002275250 | 3.167997 | AGGGAU | AGGGAU |
| RBMY1A1 | 2 | 489 | 0.003067485 | 0.0007101099 | 2.110942 | ACAAGA | ACAAGA,CAAGAC |
| SNRPB2 | 5 | 991 | 0.006134969 | 0.0014376103 | 2.093383 | AUUGCA,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM42 | 1 | 407 | 0.002044990 | 0.0005912752 | 1.790192 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 1 | 411 | 0.002044990 | 0.0005970720 | 1.776117 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| AKAP1 | 1 | 426 | 0.002044990 | 0.0006188101 | 1.724525 | AUAUAU | AUAUAU,UAUAUA |
| TUT1 | 2 | 678 | 0.003067485 | 0.0009840095 | 1.640312 | CAAUAC,CGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBFOX2 | 1 | 452 | 0.002044990 | 0.0006564894 | 1.639250 | UGCAUG | UGACUG,UGCAUG |
| HNRNPA0 | 1 | 453 | 0.002044990 | 0.0006579386 | 1.636069 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 1 | 497 | 0.002044990 | 0.0007217036 | 1.502615 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| HNRNPAB | 6 | 1782 | 0.007157464 | 0.0025839306 | 1.469881 | AAGACA,ACAAAG,AGACAA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PPRC1 | 2 | 780 | 0.003067485 | 0.0011318283 | 1.438401 | GGCGCC,GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| IGF2BP1 | 2 | 831 | 0.003067485 | 0.0012057377 | 1.347140 | AAGCAC,AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SSB | 3 | 1260 | 0.004089980 | 0.0018274462 | 1.162265 | CUGUUU,GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ACO1 | 3 | 1283 | 0.004089980 | 0.0018607779 | 1.136188 | CAGUGC,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| G3BP2 | 4 | 1644 | 0.005112474 | 0.0023839405 | 1.100673 | AGGAUA,AGGAUG,GGAUGG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| SRSF11 | 1 | 688 | 0.002044990 | 0.0009985015 | 1.034257 | AAGAAG | AAGAAG |
| RBM6 | 2 | 1054 | 0.003067485 | 0.0015289102 | 1.004552 | AUCCAA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
RBP on BSJ (Exon Only)
Plot
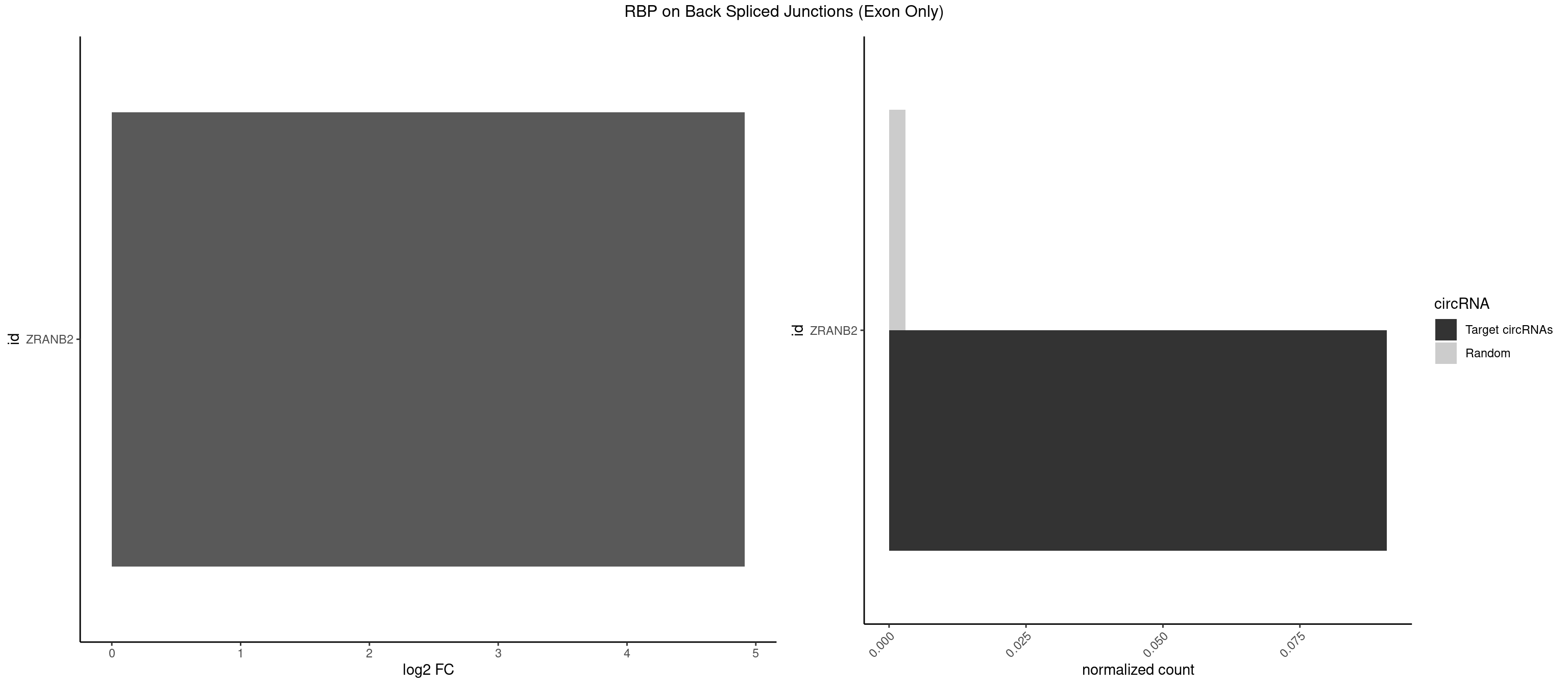
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.91523 | AAAGGU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
RBP on BSJ (Exon and Intron)
Plot
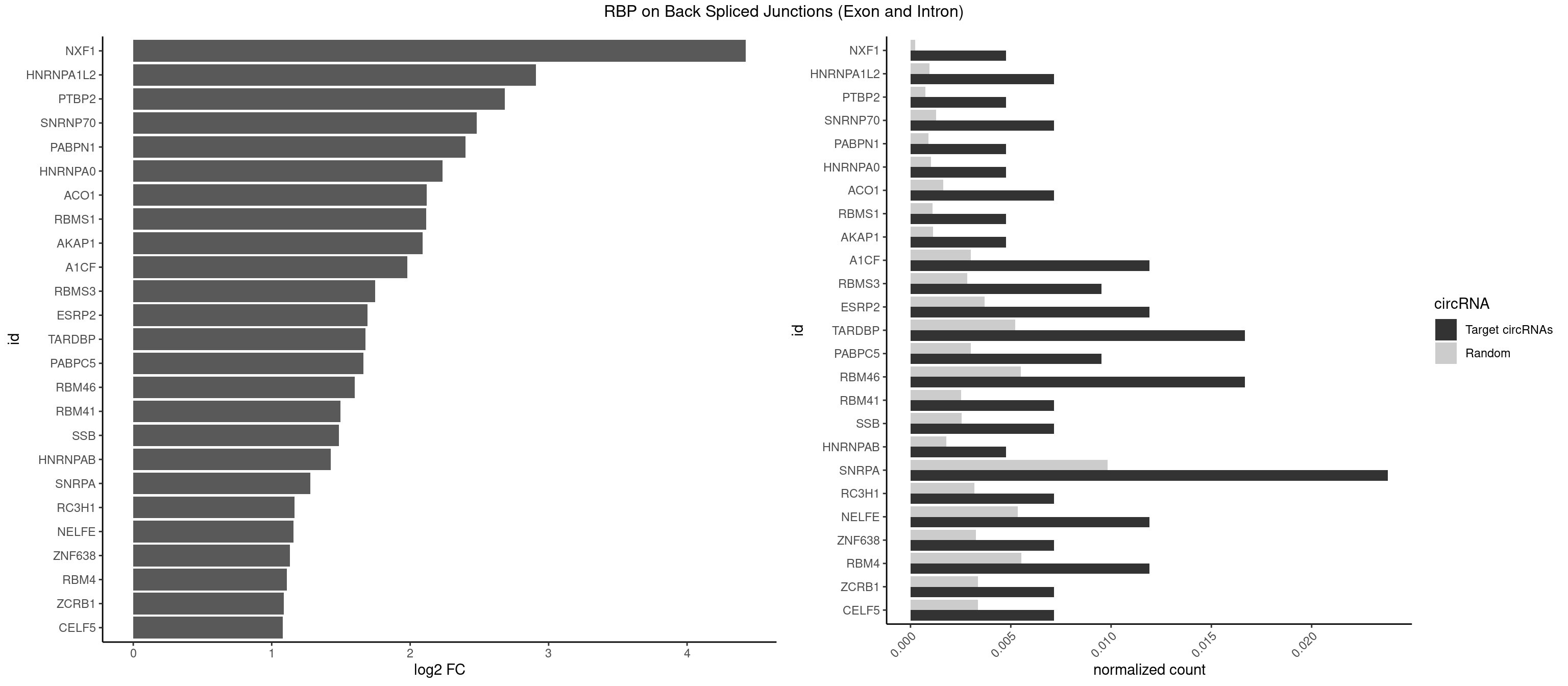
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | AUAGGG,UAGGGU | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | AAUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS1 | 1 | 217 | 0.004761905 | 0.0010983474 | 2.116204 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 1 | 221 | 0.004761905 | 0.0011185006 | 2.089973 | AUAUAU | AUAUAU,UAUAUA |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | AUAAUU,AUCAGU,UAAUUG | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAU,CUAUAU,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ESRP2 | 4 | 731 | 0.011904762 | 0.0036880290 | 1.690617 | GGGAAU,GGGGAA,UGGGGA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| TARDBP | 6 | 1034 | 0.016666667 | 0.0052146312 | 1.676328 | GAAUGA,GAAUGU,GUGAAU,UGAAUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBM46 | 6 | 1091 | 0.016666667 | 0.0055018138 | 1.598986 | AAUCAA,AAUGAA,AUCAAU,AUGAAA,AUGAAG,AUGAAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACUUU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SNRPA | 9 | 1947 | 0.023809524 | 0.0098145909 | 1.278539 | ACCUGC,AUUCCU,GAGCAG,GGUAUG,GUUUCC,UGCACA,UUGCAC,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCCUUC,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUCUCU,CUGGCU,UCUCUC | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUGGU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUUCC,CUUCCU,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,GAAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.