circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000153107:-:2:111831285:111838512
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000153107:-:2:111831285:111838512 | ENSG00000153107 | ENST00000341068 | - | 2 | 111831286 | 111838512 | 585 | UUUGACUUUGAAGGAUCACUUUCUCCUGUCAUUGCGCCCAAAAAAGCAAGGCCUUCCGAGACUGGAUCUGAUGAUGACUGGGAAUAUUUACUAAAUUCAGACUACCACCAGAAUGUUGAGUCUCAUCUUUUGAACAGAUCUUUAUGUCUGAGUCCUUCAGAAGCUUCACAGAUGAAGGAUGAGGAUUUUUCACAGAAUCUCAGUCUGGAUUCUUCUACACUUCUCUUUACUCACAUACCUGCAAUUUUUUUCGUUCUUCACCUUGUGUAUGAGGAGCUUAAGUUGAAUACUCUAAUGGGAGAAGGAAUUUGUUCACUUGUUGAACUUCUCGUUCAGUUGGCAAGGGACUUAAAAUUGGGGCCUUAUGUAGAUCAUUACUAUAGAGACUACCCAACGCUUGUCAGAACUACUGGACAAGUGUGCACAAUUGAUCCAGGUCAAACAGGAUUUAUGCAUCAUCCAUCAUUUUUUACGUCUGAGCCACCAAGUAUUUAUCAGUGGGUGAGUUCUUGUCUGAAGGGUGAAGGAAUGCCACCUUAUCCUUACCUCCCUGGAAUCUGUGAAAGAAGCAGACUUGUAGUCUUGUUUGACUUUGAAGGAUCACUUUCUCCUGUCAUUGCGCCCAAAAAAGCAAG | circ |
| ENSG00000153107:-:2:111831285:111838512 | ENSG00000153107 | ENST00000341068 | - | 2 | 111831286 | 111838512 | 22 | UUGUAGUCUUGUUUGACUUUGA | bsj |
| ENSG00000153107:-:2:111831285:111838512 | ENSG00000153107 | ENST00000341068 | - | 2 | 111838503 | 111838712 | 210 | UAUUAAUAUGAUUUCAUUUCAUCUUUACAGUAACCUGGAAGAAAGAUAUUUAAAUAAUGCCUUUCCAGAUCAAAAAAUGAGUUUUGAUGGCUUUUCUCAGUUUUCGUAAUUGGCAGAACUGAUUUCUGAAUCCUUGAAGUUGAUCAGUUGACAGGAAUGCAUUUACGAUUUUUAUCUUUUCUUUUGGGGUUACUUUUCAGUUUGACUUUG | ie_up |
| ENSG00000153107:-:2:111831285:111838512 | ENSG00000153107 | ENST00000341068 | - | 2 | 111831086 | 111831295 | 210 | UGUAGUCUUGGUAUGUAUGUGGAGUGUUACUACCUAUCUCCUUUUUCUUUUAAAAUUUAGUUUUAAAAAGUACACCUGAUAUUAAGUUUUUUUCUGUGUAUAGUUCUGUGAAUUUCAGUACGUGUAUAGAUUCAUGUAAUUACUACAAUGAGGAUAUAGAACAUUUACAUCACCCUCAAAAAUUUCUCUCAUGAUUUCGAGACUUAUCCA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
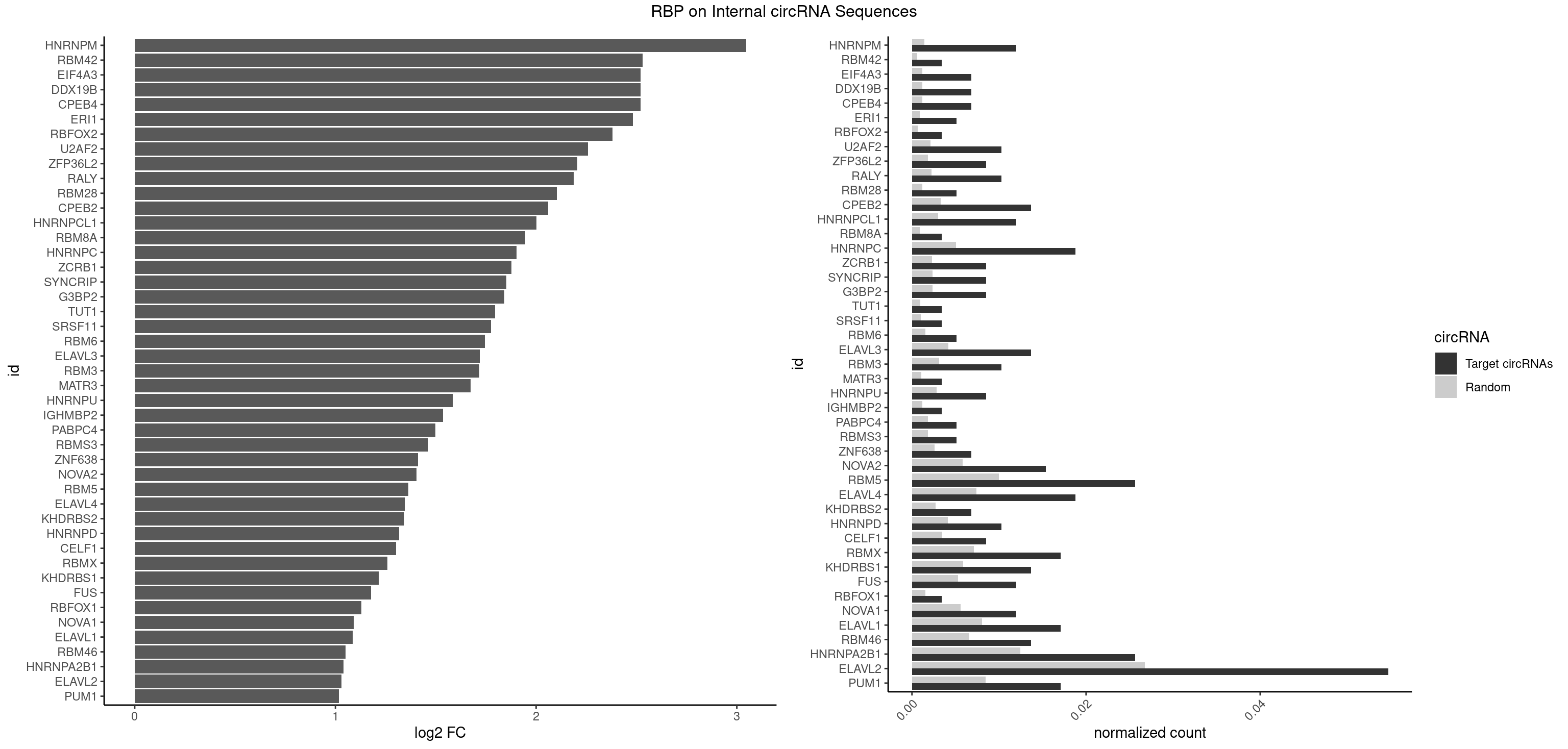
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPM | 6 | 999 | 0.011965812 | 0.0014492040 | 3.045586 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM42 | 1 | 407 | 0.003418803 | 0.0005912752 | 2.531590 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| CPEB4 | 3 | 821 | 0.006837607 | 0.0011912456 | 2.521021 | UUUUUU | UUUUUU |
| DDX19B | 3 | 821 | 0.006837607 | 0.0011912456 | 2.521021 | UUUUUU | UUUUUU |
| EIF4A3 | 3 | 821 | 0.006837607 | 0.0011912456 | 2.521021 | UUUUUU | UUUUUU |
| ERI1 | 2 | 632 | 0.005128205 | 0.0009173461 | 2.482916 | UUCAGA | UUCAGA,UUUCAG |
| RBFOX2 | 1 | 452 | 0.003418803 | 0.0006564894 | 2.380648 | UGACUG | UGACUG,UGCAUG |
| U2AF2 | 5 | 1477 | 0.010256410 | 0.0021419234 | 2.259547 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| ZFP36L2 | 4 | 1277 | 0.008547009 | 0.0018520827 | 2.206271 | AUUUAU,UAUUUA | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| RALY | 5 | 1553 | 0.010256410 | 0.0022520629 | 2.187207 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| RBM28 | 2 | 822 | 0.005128205 | 0.0011926949 | 2.104229 | UGUAGA,UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CPEB2 | 7 | 2261 | 0.013675214 | 0.0032780993 | 2.060632 | AUUUUU,CAUUUU,UUUUUU | AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| HNRNPCL1 | 6 | 2062 | 0.011965812 | 0.0029897078 | 2.000842 | AUUUUU,UUUUUU | AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| RBM8A | 1 | 611 | 0.003418803 | 0.0008869128 | 1.946627 | UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| HNRNPC | 10 | 3471 | 0.018803419 | 0.0050316361 | 1.901895 | AUUUUU,GGAUUC,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ZCRB1 | 4 | 1605 | 0.008547009 | 0.0023274215 | 1.876687 | ACUUAA,GACUUA,GAUUUA,GCUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SYNCRIP | 4 | 1634 | 0.008547009 | 0.0023694485 | 1.850868 | AAAAAA,UUUUUU | AAAAAA,UUUUUU |
| G3BP2 | 4 | 1644 | 0.008547009 | 0.0023839405 | 1.842071 | AGGAUG,AGGAUU,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| TUT1 | 1 | 678 | 0.003418803 | 0.0009840095 | 1.796747 | AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SRSF11 | 1 | 688 | 0.003418803 | 0.0009985015 | 1.775655 | AAGAAG | AAGAAG |
| RBM6 | 2 | 1054 | 0.005128205 | 0.0015289102 | 1.745950 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ELAVL3 | 7 | 2867 | 0.013675214 | 0.0041563169 | 1.718186 | AUUUAU,UAUUUA,UUUUUU | AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| RBM3 | 5 | 2152 | 0.010256410 | 0.0031201361 | 1.716845 | AAUACU,AGACUA,GAGACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| MATR3 | 1 | 739 | 0.003418803 | 0.0010724109 | 1.672634 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| HNRNPU | 4 | 1965 | 0.008547009 | 0.0028491350 | 1.584896 | AAAAAA,UUUUUU | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| IGHMBP2 | 1 | 813 | 0.003418803 | 0.0011796520 | 1.535130 | AAAAAA | AAAAAA |
| PABPC4 | 2 | 1251 | 0.005128205 | 0.0018144033 | 1.498959 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBMS3 | 2 | 1283 | 0.005128205 | 0.0018607779 | 1.462548 | CUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| ZNF638 | 3 | 1773 | 0.006837607 | 0.0025708878 | 1.411225 | CGUUCU,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| NOVA2 | 8 | 4013 | 0.015384615 | 0.0058171047 | 1.403115 | ACCACC,AGAUCA,AUCAUC,GAUCAC,UAGAUC,UUUUUU | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| RBM5 | 14 | 6879 | 0.025641026 | 0.0099705232 | 1.362713 | AAAAAA,AAGGAA,AGGGUG,CAAGGG,CUUCUC,GAAGGA,GAAGGG,UCUUCU,UUCUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| ELAVL4 | 10 | 5105 | 0.018803419 | 0.0073996354 | 1.345469 | AAAAAA,AUUUAU,UAUUUA,UCUAAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| KHDRBS2 | 3 | 1858 | 0.006837607 | 0.0026940701 | 1.343704 | UUUUUU | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| HNRNPD | 5 | 2837 | 0.010256410 | 0.0041128408 | 1.318319 | AAAAAA,AUUUAU,UAUUUA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| CELF1 | 4 | 2391 | 0.008547009 | 0.0034664959 | 1.301942 | UGUCUG,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| RBMX | 9 | 4925 | 0.017094017 | 0.0071387787 | 1.259742 | AAGAAG,AAGGAA,AAGUGU,AGAAGG,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| KHDRBS1 | 7 | 4064 | 0.013675214 | 0.0058910141 | 1.214975 | AUUUAC,CUAAAU,UUAAAA,UUUUAC,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| FUS | 6 | 3648 | 0.011965812 | 0.0052881452 | 1.178085 | AAAAAA,GGGUGA,UUUUUU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| RBFOX1 | 1 | 1077 | 0.003418803 | 0.0015622419 | 1.129874 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| NOVA1 | 6 | 3872 | 0.011965812 | 0.0056127669 | 1.092134 | ACCACC,AUCAUC,UCAGUC,UUUUUU | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| ELAVL1 | 9 | 5554 | 0.017094017 | 0.0080503280 | 1.086372 | AUUUAU,UAUUUA,UUGUUU,UUUGUU,UUUUUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| RBM46 | 7 | 4554 | 0.013675214 | 0.0066011240 | 1.050780 | AUCAUU,AUGAAG,AUGAUG,GAUCAU,GAUGAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPA2B1 | 14 | 8607 | 0.025641026 | 0.0124747476 | 1.039443 | AAGAAG,AAGGAA,AGAAGC,AGACUA,AGGAGC,CAAGGG,GAAGGA,GGGGCC | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| ELAVL2 | 31 | 18468 | 0.054700855 | 0.0267653478 | 1.031197 | AAUUUG,AAUUUU,AUAUUU,AUUUAC,AUUUAU,AUUUUU,CAUUUU,CUUUCU,GAUUUA,GAUUUU,UAAGUU,UAUUUA,UCAUUU,UCUUUU,UUAAGU,UUAUGU,UUUACU,UUUAUG,UUUUUA,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PUM1 | 9 | 5823 | 0.017094017 | 0.0084401638 | 1.018149 | AAUAUU,AAUGUU,CAGAAU,CCAGAA,CUUGUA,GUAGAU,UGUAGA,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
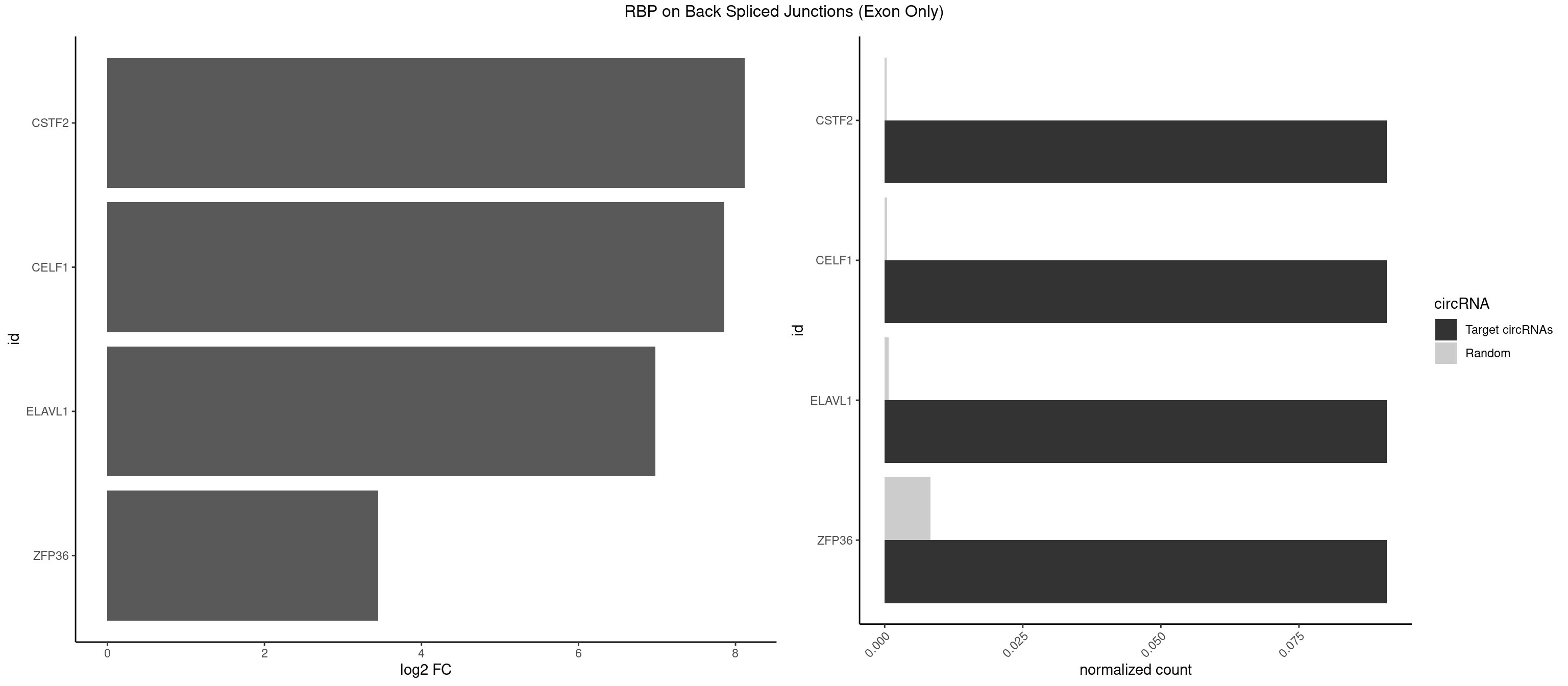
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| CSTF2 | 1 | 4 | 0.09090909 | 0.0003274823 | 8.116864 | UGUUUG | GUGUUG,GUUUUG,UGUGUG,UGUUUG |
| CELF1 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | UGUUUG | GUGUUG,UGUCUG,UGUGUG,UGUUGU,UGUUUG |
| ELAVL1 | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | UUGUUU | UAUUUU,UGGUUU,UUAGUU,UUGGUU,UUUAGU,UUUGGU,UUUGUU |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | UCUUGU | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
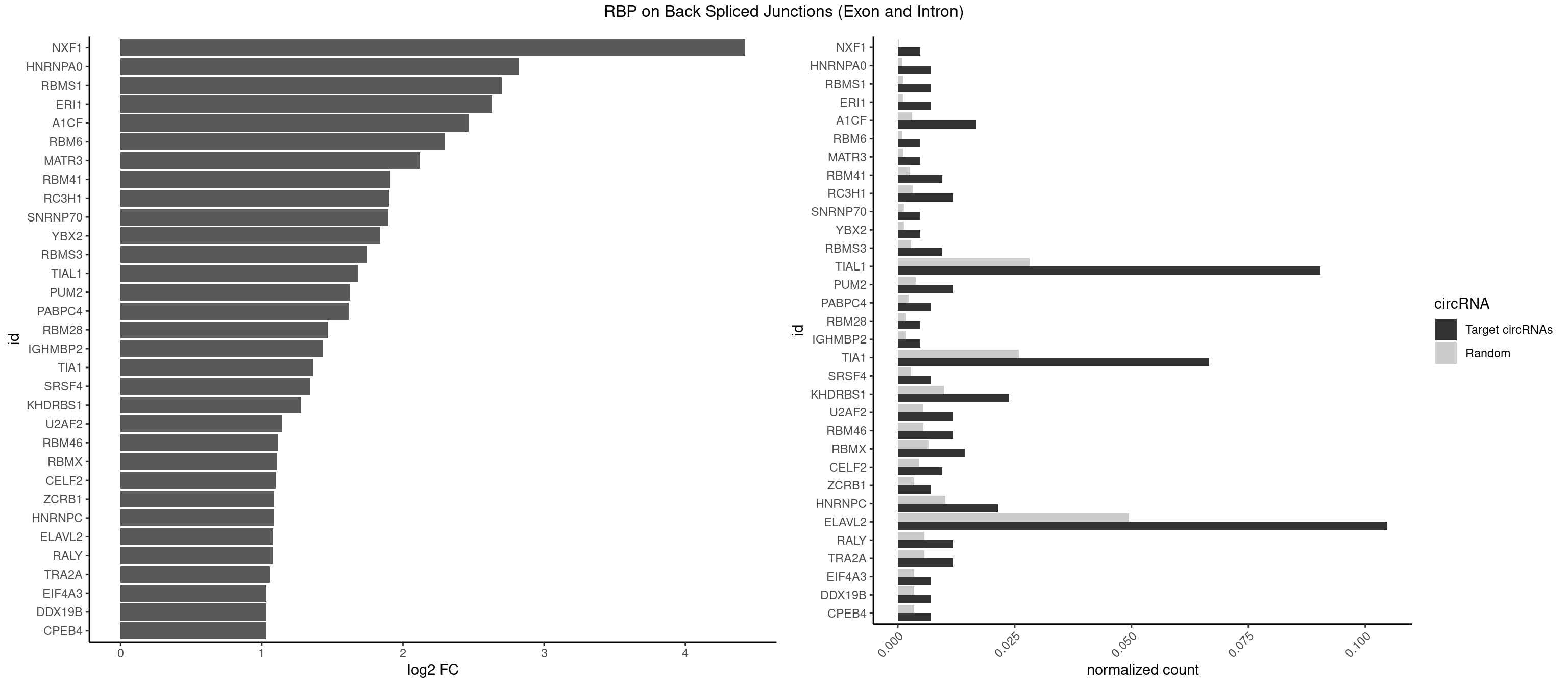
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| HNRNPA0 | 2 | 200 | 0.007142857 | 0.0010126965 | 2.818299 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMS1 | 2 | 217 | 0.007142857 | 0.0010983474 | 2.701167 | AUAUAG,GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUUCAG | UUCAGA,UUUCAG |
| A1CF | 6 | 598 | 0.016666667 | 0.0030179363 | 2.465331 | AUCAGU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM6 | 1 | 191 | 0.004761905 | 0.0009673519 | 2.299426 | UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM41 | 3 | 502 | 0.009523810 | 0.0025342604 | 1.909974 | UACUUU,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RC3H1 | 4 | 631 | 0.011904762 | 0.0031841999 | 1.902536 | UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| RBMS3 | 3 | 562 | 0.009523810 | 0.0028365578 | 1.747397 | AUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| TIAL1 | 37 | 5597 | 0.090476190 | 0.0282043531 | 1.681620 | AAAUUU,AGUUUU,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UCAGUU,UUAAAU,UUCAGU,UUUAAA,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | UAAAUA,UACAUC,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| TIA1 | 27 | 5140 | 0.066666667 | 0.0259018541 | 1.363910 | AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UCCUUU,UCUCCU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUUA,UUUUUC,UUUUUU | AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| KHDRBS1 | 9 | 1945 | 0.023809524 | 0.0098045143 | 1.280021 | AUUUAA,AUUUAC,UAAAAA,UUAAAA,UUAAAU,UUUUUU | AUAAAA,AUUUAA,AUUUAC,CAAAAU,CUAAAA,CUAAAU,GAAAAC,UAAAAA,UAAAAC,UAAAAG,UUAAAA,UUAAAU,UUUUAC,UUUUUU |
| U2AF2 | 4 | 1071 | 0.011904762 | 0.0054010480 | 1.140228 | UUUUUC,UUUUUU | UUUUCC,UUUUUC,UUUUUU |
| RBM46 | 4 | 1091 | 0.011904762 | 0.0055018138 | 1.113560 | AUCAAA,AUGAUU,GAUCAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBMX | 5 | 1316 | 0.014285714 | 0.0066354293 | 1.106311 | AGUAAC,AGUGUU,AUCAAA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | GUAUGU,UAUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AUUUAA,GACUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| HNRNPC | 8 | 2006 | 0.021428571 | 0.0101118501 | 1.083489 | AUUUUU,CUUUUU,GGAUAU,UUUUUA,UUUUUC,UUUUUU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL2 | 43 | 9826 | 0.104761905 | 0.0495112858 | 1.081285 | AAUUUA,AUAUUU,AUUUAA,AUUUAC,AUUUAG,AUUUUU,CUUUUA,CUUUUU,GAUUUU,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UACUUU,UAUUUA,UCAUUU,UCUUUU,UGAUUU,UUAAGU,UUACUU,UUAGUU,UUCAUU,UUCUUU,UUUAGU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAU,UUUUUA,UUUUUC,UUUUUU | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| RALY | 4 | 1117 | 0.011904762 | 0.0056328094 | 1.079612 | UUUUUC,UUUUUU | UUUUUC,UUUUUG,UUUUUU |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAGAAA,AGAAAG,GAAAGA,GAAGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.