circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000158019:+:2:28025225:28129380
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000158019:+:2:28025225:28129380 | ENSG00000158019 | ENST00000344773 | + | 2 | 28025226 | 28129380 | 380 | AAUCUUGCCUCCUGGAAUCCUUCAAAUCCUGAAUGUCUCUUACUUGUGGUGAAGGAACUUGUGCAACAAUAUCACCAAUUCCAAUGUAGCCGCCUCCGGGAGAGCUCCCGCCUCAUGUUUGAAUACCAGACAUUACUGGAGGAGCCACAGUAUGGAGAGAACAUGGAAAUUUAUGCUGGGAAAAAAAACAACUGGACUGGUGAAUUUUCAGCUCGUUUCCUUUUGAAGCUGCCCGUAGAUUUCAGCAAUAUCCCCACAUACCUUCUCAAGGAUGUAAAUGAAGACCCUGGAGAAGAUGUGGCCCUCCUCUCUGUUAGUUUUGAGGACACUGAAGCCACCCAGGUGUACCCCAAGCUGUACUUGUCACCUCGAAUUGAGCAAAUCUUGCCUCCUGGAAUCCUUCAAAUCCUGAAUGUCUCUUACUUGUGGU | circ |
| ENSG00000158019:+:2:28025225:28129380 | ENSG00000158019 | ENST00000344773 | + | 2 | 28025226 | 28129380 | 22 | CGAAUUGAGCAAAUCUUGCCUC | bsj |
| ENSG00000158019:+:2:28025225:28129380 | ENSG00000158019 | ENST00000344773 | + | 2 | 28025026 | 28025235 | 210 | GAUUUUUUUAGAAGGCAGGAGAAAUUGCCAAUAUCAAAAGAUUUCUCUAACUUCAUCUCUGGGUAUAUGGCUAAGGAGGAGUAGUCUGUUCCUCUUAGAUUUCUAUUACUCUUUUCCUCUACAUUGAUGUGUGAAUGUGAUACUGGAUUUGUAUGAAAUUUCAGUUUUAACUGGUCUUUUAUUUGUUACGACUUCUACAGAAUCUUGCCU | ie_up |
| ENSG00000158019:+:2:28025225:28129380 | ENSG00000158019 | ENST00000344773 | + | 2 | 28129371 | 28129580 | 210 | GAAUUGAGCAGUAAGUGUCAUUAACAUGAGGUAGCCUGGUGCUACUUCUUGCUAACCAAUAUGUCUCAUUCUUUCUUUUGCCACUCUUGAACUCUUACAUUUAUUUCUCUUCAUUGACUUUUGUUUUCUUAAAAAAUUCAUCAUAUUAAGAGUUUCCAUUGCAGAGUAAAUGCAGUUAGUACUUGCCACUACACCCUUUUGGCAAGUGUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
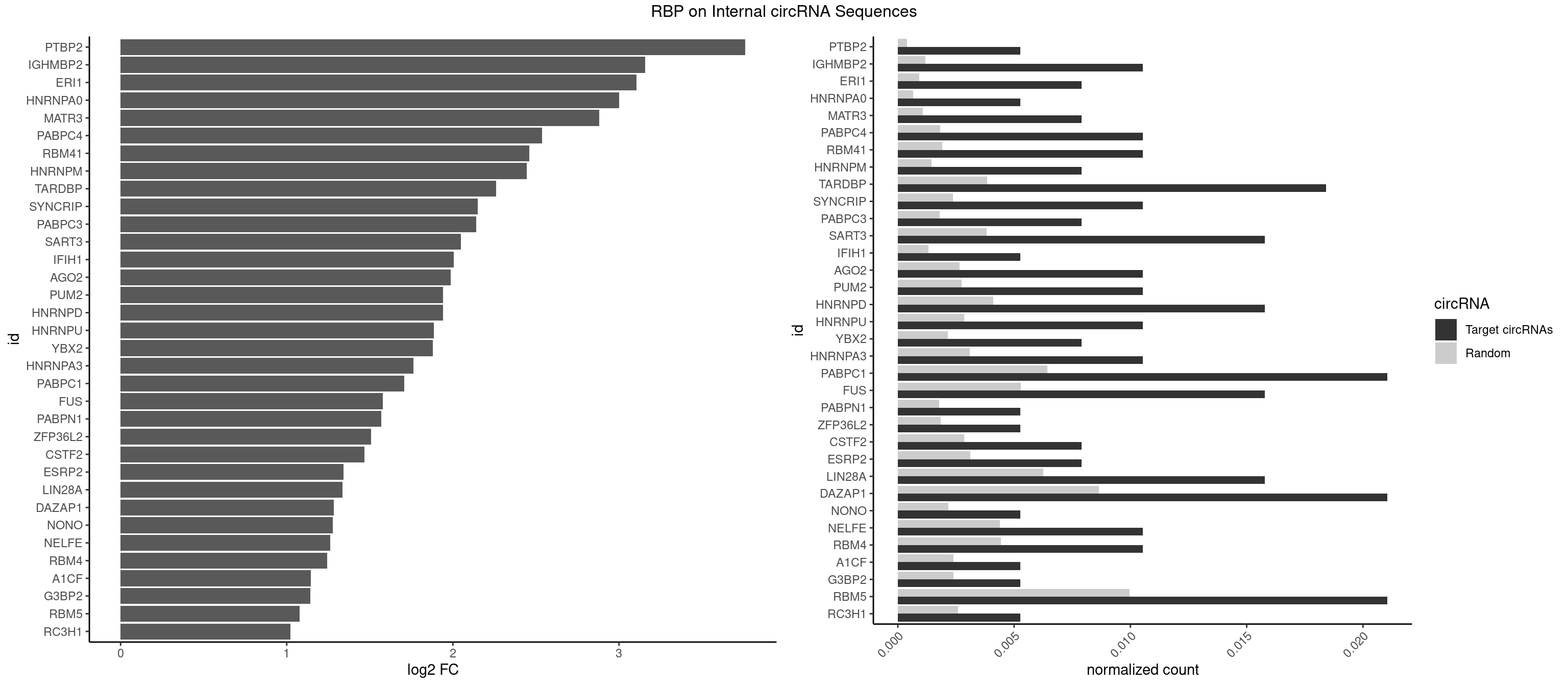
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 267 | 0.005263158 | 0.0003883867 | 3.760363 | CUCUCU | CUCUCU |
| IGHMBP2 | 3 | 813 | 0.010526316 | 0.0011796520 | 3.157567 | AAAAAA | AAAAAA |
| ERI1 | 2 | 632 | 0.007894737 | 0.0009173461 | 3.105353 | UUUCAG | UUCAGA,UUUCAG |
| HNRNPA0 | 1 | 453 | 0.005263158 | 0.0006579386 | 2.999904 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| MATR3 | 2 | 739 | 0.007894737 | 0.0010724109 | 2.880033 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PABPC4 | 3 | 1251 | 0.010526316 | 0.0018144033 | 2.536433 | AAAAAA | AAAAAA,AAAAAG |
| RBM41 | 3 | 1318 | 0.010526316 | 0.0019115000 | 2.461223 | UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| HNRNPM | 2 | 999 | 0.007894737 | 0.0014492040 | 2.445631 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| TARDBP | 6 | 2654 | 0.018421053 | 0.0038476365 | 2.259311 | GAAUGU,GUGAAU,GUUUUG,UGAAUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SYNCRIP | 3 | 1634 | 0.010526316 | 0.0023694485 | 2.151377 | AAAAAA | AAAAAA,UUUUUU |
| PABPC3 | 2 | 1234 | 0.007894737 | 0.0017897669 | 2.141119 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SART3 | 5 | 2634 | 0.015789474 | 0.0038186524 | 2.047828 | AAAAAA,AAAAAC,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| IFIH1 | 1 | 904 | 0.005263158 | 0.0013115296 | 2.004678 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| AGO2 | 3 | 1830 | 0.010526316 | 0.0026534924 | 1.988036 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PUM2 | 3 | 1890 | 0.010526316 | 0.0027404447 | 1.941519 | GUAAAU,GUAGAU,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| HNRNPD | 5 | 2837 | 0.015789474 | 0.0041128408 | 1.940756 | AAAAAA,AAUUUA,AUUUAU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| HNRNPU | 3 | 1965 | 0.010526316 | 0.0028491350 | 1.885405 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| YBX2 | 2 | 1480 | 0.007894737 | 0.0021462711 | 1.879059 | AACAAC,ACAACU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| HNRNPA3 | 3 | 2140 | 0.010526316 | 0.0031027457 | 1.762383 | AGGAGC,CAAGGA,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| PABPC1 | 7 | 4443 | 0.021052632 | 0.0064402624 | 1.708809 | AAAAAA,AAAAAC,CAAAUC,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| FUS | 5 | 3648 | 0.015789474 | 0.0052881452 | 1.578129 | AAAAAA,UGGUGA | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| PABPN1 | 1 | 1222 | 0.005263158 | 0.0017723764 | 1.570244 | AGAAGA | AAAAGA,AGAAGA |
| ZFP36L2 | 1 | 1277 | 0.005263158 | 0.0018520827 | 1.506780 | AUUUAU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CSTF2 | 2 | 1967 | 0.007894737 | 0.0028520334 | 1.468900 | GUUUUG,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ESRP2 | 2 | 2150 | 0.007894737 | 0.0031172377 | 1.340623 | GGGAAA,UGGGAA | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| LIN28A | 5 | 4315 | 0.015789474 | 0.0062547643 | 1.335936 | GGAGAA,GGAGGA,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| DAZAP1 | 7 | 5964 | 0.021052632 | 0.0086445016 | 1.284146 | AAAAAA,AAUUUA,AGGAUG,AGUAUG,UAGUUU | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| NONO | 1 | 1498 | 0.005263158 | 0.0021723567 | 1.276668 | AGGAAC | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| NELFE | 3 | 3028 | 0.010526316 | 0.0043896388 | 1.261826 | CUCUCU,GUCUCU,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM4 | 3 | 3065 | 0.010526316 | 0.0044432593 | 1.244310 | CCUUCU,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| A1CF | 1 | 1642 | 0.005263158 | 0.0023810421 | 1.144336 | CAGUAU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| G3BP2 | 1 | 1644 | 0.005263158 | 0.0023839405 | 1.142580 | AGGAUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM5 | 7 | 6879 | 0.021052632 | 0.0099705232 | 1.078259 | AAAAAA,AAGGAA,CAAGGA,CUUCUC,GAAGGA | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| RC3H1 | 1 | 1786 | 0.005263158 | 0.0025897275 | 1.023128 | CCUUCU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
RBP on BSJ (Exon Only)
Plot
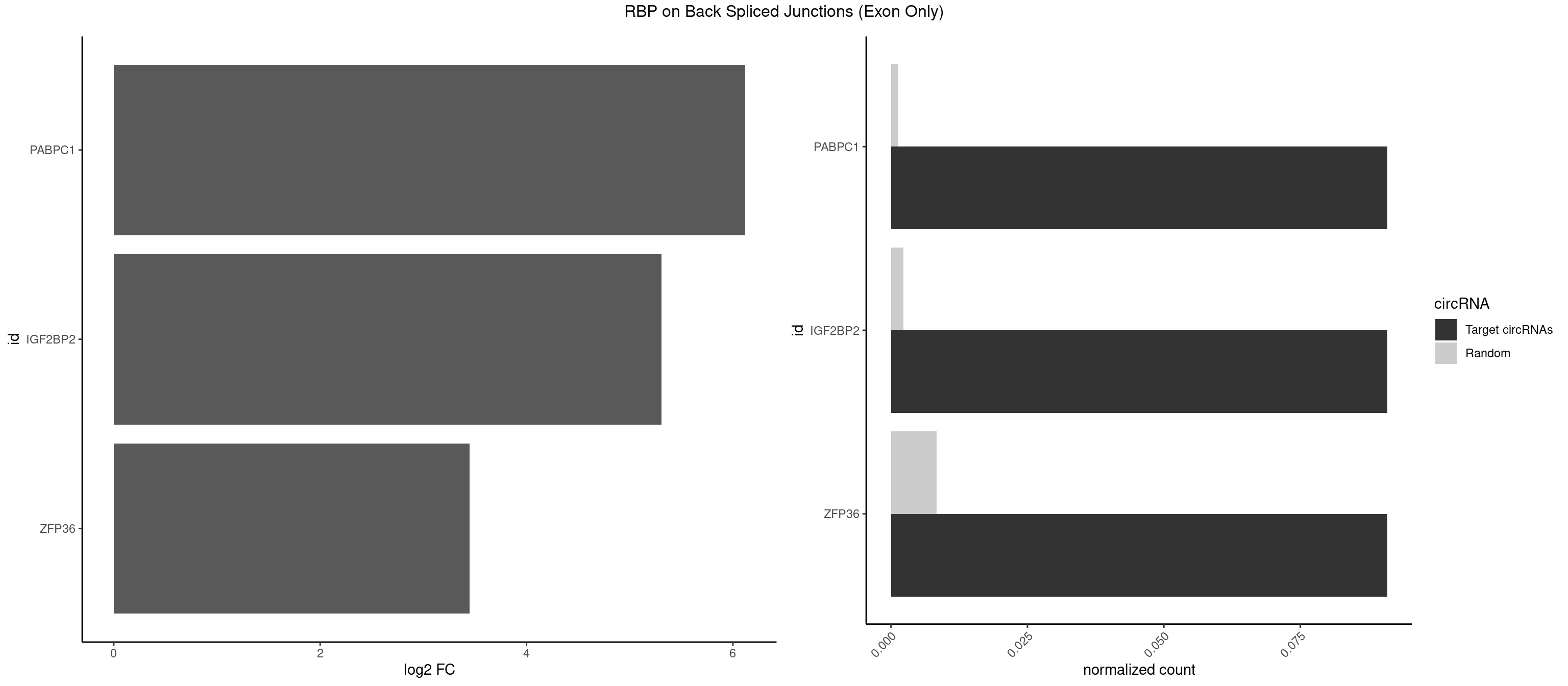
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PABPC1 | 1 | 19 | 0.09090909 | 0.001309929 | 6.116864 | CAAAUC | AAAAAA,ACAAAC,ACGAAU,AGAAAA,CAAACC,CAAAUA,CGAACA,CUAAUA,GAAAAA,GAAAAC |
| IGF2BP2 | 1 | 34 | 0.09090909 | 0.002292376 | 5.309509 | CAAAUC | AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,ACAAAC,ACAAUC,ACACUC,ACAUAC,CAAAAC,CAAUAC,CAAUCA,CAAUUC,CACUCA,CAUACA,CAUUCA,CCAUAC,CCAUUC,GAAAAC,GAACAC,GAAUAC,GAAUUC |
| ZFP36 | 1 | 126 | 0.09090909 | 0.008318051 | 3.450107 | AGCAAA | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
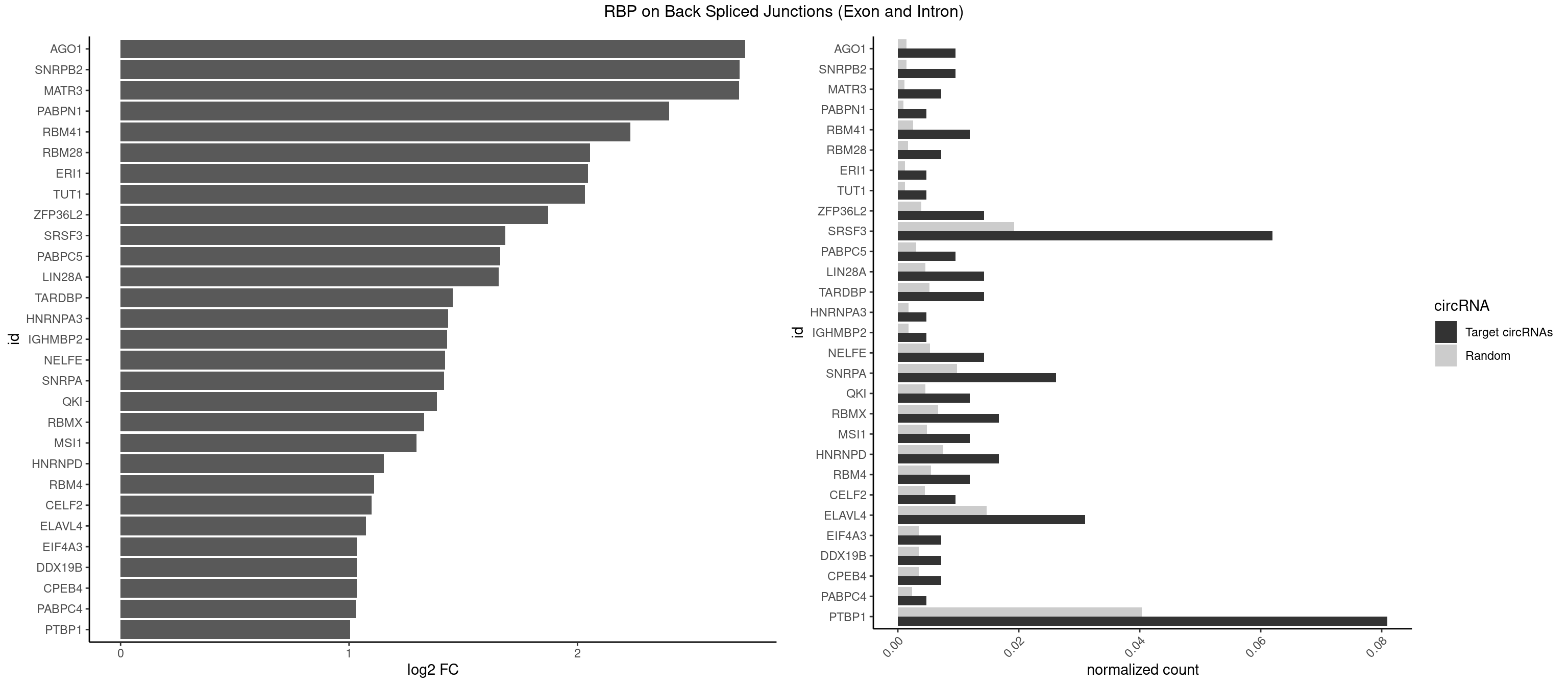
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| AGO1 | 3 | 283 | 0.009523810 | 0.0014308746 | 2.734641 | AGGUAG,GAGGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SNRPB2 | 3 | 288 | 0.009523810 | 0.0014560661 | 2.709463 | AUUGCA,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PABPN1 | 1 | 178 | 0.004761905 | 0.0009018541 | 2.400573 | AAAAGA | AAAAGA,AGAAGA |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | UACAUU,UACUUG,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGU,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.0011638452 | 2.032640 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| ZFP36L2 | 5 | 774 | 0.014285714 | 0.0039046755 | 1.871299 | AUUUAU,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| SRSF3 | 25 | 3825 | 0.061904762 | 0.0192765014 | 1.683207 | ACGACU,ACUACA,ACUUCA,ACUUCU,AUUCAU,CACUAC,CAUCAU,CCUCUU,CUACAC,CUACAG,CUCUUC,CUUCAU,CUUCUU,GACUUC,UACGAC,UACUUC,UAUCAA,UCAUCA,UCAUCU,UCUUCA,UUACGA,UUCAUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| PABPC5 | 3 | 596 | 0.009523810 | 0.0030078597 | 1.662801 | AGAAAU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| LIN28A | 5 | 900 | 0.014285714 | 0.0045395002 | 1.653968 | AGGAGA,AGGAGU,GGAGAA,GGAGGA,GGAGUA | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| TARDBP | 5 | 1034 | 0.014285714 | 0.0052146312 | 1.453936 | GAAUGU,GUGAAU,UGAAUG,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| HNRNPA3 | 1 | 349 | 0.004761905 | 0.0017634019 | 1.433177 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| NELFE | 5 | 1059 | 0.014285714 | 0.0053405885 | 1.419503 | CUCUGG,GCUAAC,GGCUAA,UCUCUG,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SNRPA | 10 | 1947 | 0.026190476 | 0.0098145909 | 1.416042 | AGCAGU,AGGAGA,AGUAGU,AUUGCA,GAGCAG,GCAGUA,GGGUAU,GUAGUC,GUUUCC,UUUCCU | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| QKI | 4 | 904 | 0.011904762 | 0.0045596534 | 1.384543 | AUCAUA,AUUAAC,CUAACC,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBMX | 6 | 1316 | 0.016666667 | 0.0066354293 | 1.328704 | AAGUGU,AGAAGG,AGUGUU,AUCAAA,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGAGG,AGGUAG,AGUAAG,AGUUAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| HNRNPD | 6 | 1488 | 0.016666667 | 0.0075020153 | 1.151615 | AAAAAA,AUUUAU,UUAUUU,UUUAUU | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUCUU,CUCUUU,CUUCUU,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| CELF2 | 3 | 881 | 0.009523810 | 0.0044437727 | 1.099754 | AUGUGU,GUCUGU,UGUGUG | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ELAVL4 | 12 | 2916 | 0.030952381 | 0.0146966949 | 1.074559 | AAAAAA,AUUUAU,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| CPEB4 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| DDX19B | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| EIF4A3 | 2 | 691 | 0.007142857 | 0.0034864974 | 1.034723 | UUUUUU | UUUUUU |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAA | AAAAAA,AAAAAG |
| PTBP1 | 33 | 8003 | 0.080952381 | 0.0403264813 | 1.005346 | ACUUUU,AUUUUU,CCUCUU,CCUUUU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCUU,CUUUCU,CUUUUC,GUCUUU,UCCUCU,UCUCUU,UCUUUC,UCUUUU,UUAUUU,UUCUCU,UUCUUG,UUCUUU,UUUCUU,UUUUCC,UUUUCU,UUUUUU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.