circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000184465_ENSG00000272848:-:6:169632946:169638660
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000184465_ENSG00000272848:-:6:169632946:169638660 | ENSG00000184465_ENSG00000272848 | MSTRG.29337.7 | - | 6 | 169632947 | 169638660 | 342 | GUCACGACGGGGCAGUGAAUGCCGUGUGCUGGAGCCAGGACCGGAGGUGGCUGCUCUCUGCGGCCCGGGACGGGACCCUGCGAAUGUGGUCGGCUCGUGGGGCAGAGCUCGCACUGCUUCUGAUAUAAACAGAAGAGCAAGUCCAAGCUGAUUUGCAGGCUCUCCACGACGGGUGCAGUAGACAUGACCAGUUUAUCGGCAGUCAACGACUUUUAUUCCCACAUCGUACUCGCAGCUGGCCGGAACAGGACCGUGGAAGUGUUUGACCUCAACGCCGGCUGCAGUGCAGCGGUGAUAGCGGAAGCCCACUCACGGCCUGUCCAUCAAAUCUGCCAAAAUAAAGUCACGACGGGGCAGUGAAUGCCGUGUGCUGGAGCCAGGACCGGAGGUGG | circ |
| ENSG00000184465_ENSG00000272848:-:6:169632946:169638660 | ENSG00000184465_ENSG00000272848 | MSTRG.29337.7 | - | 6 | 169632947 | 169638660 | 22 | GCCAAAAUAAAGUCACGACGGG | bsj |
| ENSG00000184465_ENSG00000272848:-:6:169632946:169638660 | ENSG00000184465_ENSG00000272848 | MSTRG.29337.7 | - | 6 | 169638651 | 169638860 | 210 | AGUCUCUACUGAAGGGAGAUGGUGCACCCUGCCUCAGGCUCUGCAGCCUCACUGUGGCGAGGAGAAGCCUCCUGGCGCGCCCCUAAUUACUUGAAAAAUGCUGUUCCAGUUGUGUUGUAUGAAAGUACCAGAUAAGUAGCACUCAGUCAAAACUGAUAUUAAUAGUAGAAAUAGUAACCUUCUGUGGUCAUUCCUGUUAGGUCACGACGG | ie_up |
| ENSG00000184465_ENSG00000272848:-:6:169632946:169638660 | ENSG00000184465_ENSG00000272848 | MSTRG.29337.7 | - | 6 | 169632747 | 169632956 | 210 | CCAAAAUAAAGUGAGUAAGUUUUUGGAUAACAUGUAUCAUCUGUAAAACUAUGUGCUUAAAGCAUUUAACAGAUACAAAUGUAUGAUGCCAUUUUAUAAGUGUUUGCUACCAAAUUCAAUGUUAAUUUUUAUUGAUCAUUAAAUUAUUCGAAGCCUUGAUGUUUGACCCUAGGAGAUAUUUUAAUAUCAUGUUGAAUGUUAAGUUUUCUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
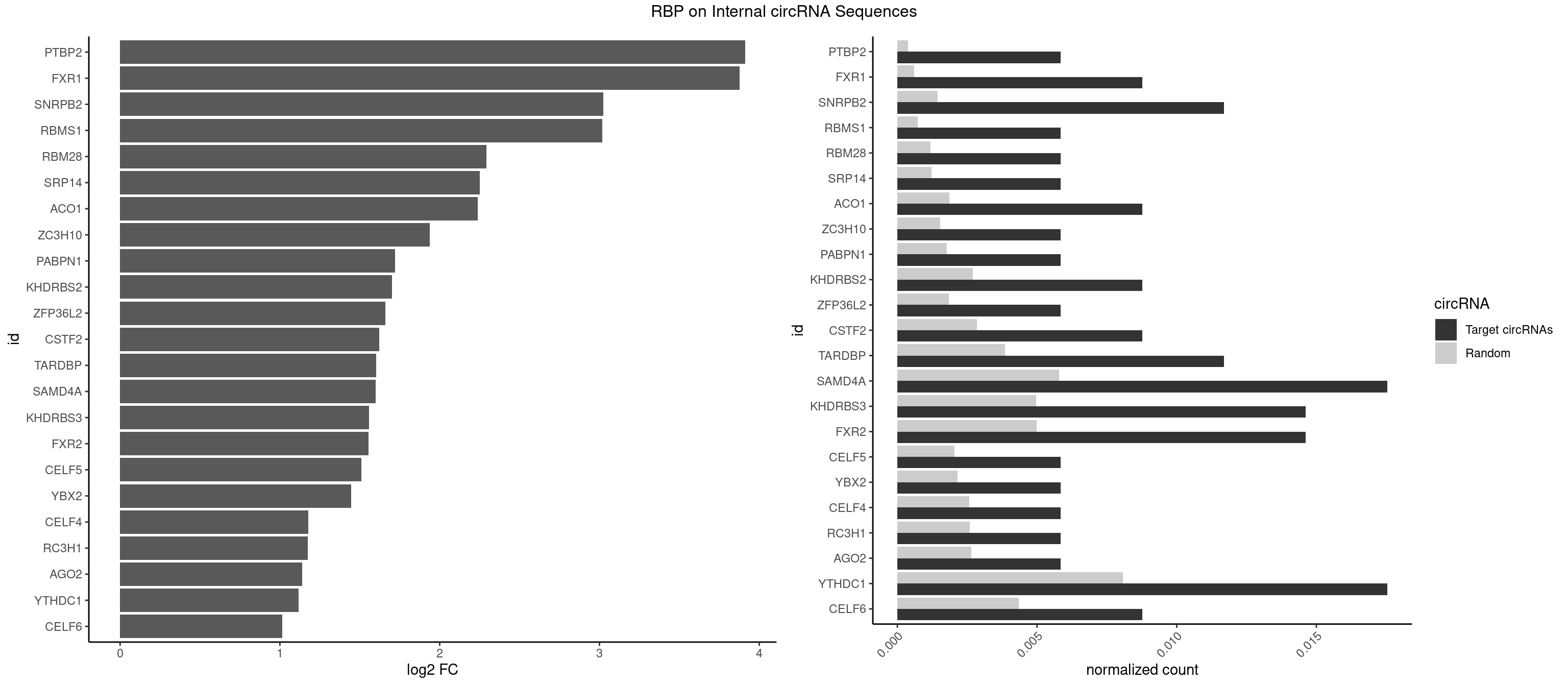
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 267 | 0.005847953 | 0.0003883867 | 3.912366 | CUCUCU | CUCUCU |
| FXR1 | 2 | 411 | 0.008771930 | 0.0005970720 | 3.876917 | ACGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| SNRPB2 | 3 | 991 | 0.011695906 | 0.0014376103 | 3.024259 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBMS1 | 1 | 497 | 0.005847953 | 0.0007217036 | 3.018453 | GAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBM28 | 1 | 822 | 0.005847953 | 0.0011926949 | 2.293707 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SRP14 | 1 | 847 | 0.005847953 | 0.0012289250 | 2.250535 | GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| ACO1 | 2 | 1283 | 0.008771930 | 0.0018607779 | 2.236988 | CAGUGA,CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| ZC3H10 | 1 | 1053 | 0.005847953 | 0.0015274610 | 1.936796 | GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PABPN1 | 1 | 1222 | 0.005847953 | 0.0017723764 | 1.722247 | AGAAGA | AAAAGA,AGAAGA |
| KHDRBS2 | 2 | 1858 | 0.008771930 | 0.0026940701 | 1.703107 | AAUAAA,AUAAAC | AAUAAA,AUAAAA,AUAAAC,GAUAAA,UUUUUU |
| ZFP36L2 | 1 | 1277 | 0.005847953 | 0.0018520827 | 1.658783 | UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| CSTF2 | 2 | 1967 | 0.008771930 | 0.0028520334 | 1.620903 | GUGUUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 3 | 2654 | 0.011695906 | 0.0038476365 | 1.603959 | GAAUGU,GUGAAU,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SAMD4A | 5 | 3992 | 0.017543860 | 0.0057866714 | 1.600161 | CGGGAC,CUGGCC,GCUGGA,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| KHDRBS3 | 4 | 3429 | 0.014619883 | 0.0049707696 | 1.556391 | AAAUAA,AAUAAA,AUAAAC,AUAAAG | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
| FXR2 | 4 | 3434 | 0.014619883 | 0.0049780156 | 1.554289 | GACGGG,GGACGG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| CELF5 | 1 | 1415 | 0.005847953 | 0.0020520728 | 1.510850 | GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| YBX2 | 1 | 1480 | 0.005847953 | 0.0021462711 | 1.446099 | ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF4 | 1 | 1782 | 0.005847953 | 0.0025839306 | 1.178364 | GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RC3H1 | 1 | 1786 | 0.005847953 | 0.0025897275 | 1.175131 | CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| AGO2 | 1 | 1830 | 0.005847953 | 0.0026534924 | 1.140039 | UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| YTHDC1 | 5 | 5576 | 0.017543860 | 0.0080822104 | 1.118144 | GAAUGC,GGCUGC,GGGUGC,UCGUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| CELF6 | 2 | 2997 | 0.008771930 | 0.0043447134 | 1.013633 | GUGGGG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
RBP on BSJ (Exon Only)
Plot
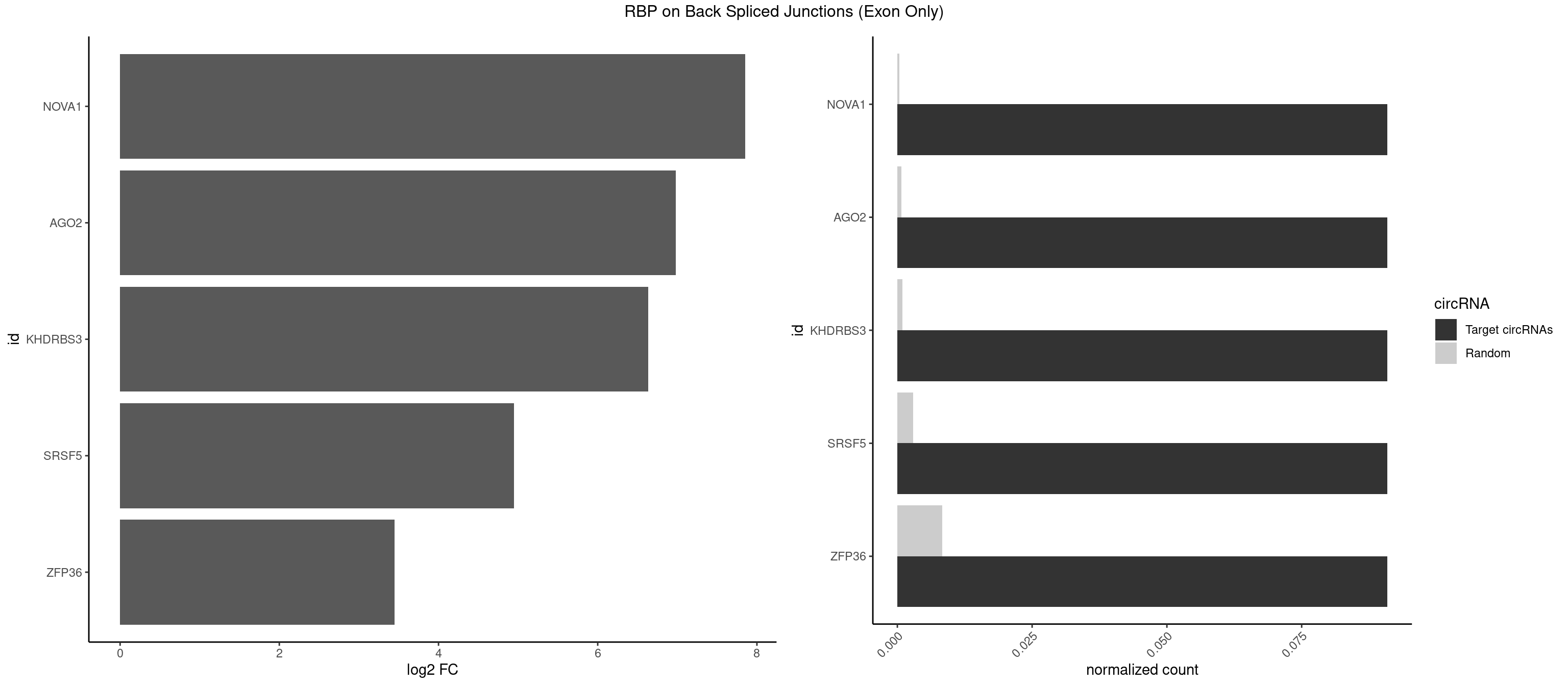
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NOVA1 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | AGUCAC | ACCACC,CAGUCA,CAUUCA,UCAGUC |
| AGO2 | 1 | 10 | 0.09090909 | 0.0007204611 | 6.979360 | UAAAGU | AAAAAA,AAAGUG,UAAAGU |
| KHDRBS3 | 1 | 13 | 0.09090909 | 0.0009169505 | 6.631437 | AUAAAG | AAAUAA,AGAUAA,AUAAAG,AUUAAA,GAUAAA,UAAAUA,UUAAAC |
| SRSF5 | 1 | 44 | 0.09090909 | 0.0029473408 | 4.946939 | AUAAAG | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUC,CACGGA,CGCAGC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACGUA,UGCAGA,UGCAGC,UGCGGC |
| ZFP36 | 1 | 126 | 0.09090909 | 0.0083180508 | 3.450107 | AUAAAG | AAAAAA,AAAAAG,AAAAAU,AAAAGA,AAAAGC,AAAAGG,AAAAGU,AAAAUA,AAAGAA,AAAGCA,AAAUAA,AACAAA,AACAAG,AAGAAA,AAGCAA,AAGGAA,AAGUAA,AAUAAG,ACAAAC,ACAAAG,ACAAGC,ACAAGG,ACAAGU,ACCUGU,AGAAAG,AGGAAA,AGUAAA,AUAAAG,AUAAAU,AUAAGC,AUUUAA,CAAAGA,CAAAUA,CAAGGA,CAAGUA,CCCUCC,CCCUGU,CCUUCU,GCAAAG,GGAAAG,GUAAAG,UAAAGA,UAAAUA,UAAGCA,UAAGGA,UCCUCU,UCCUGC,UCCUGU,UCUUCC,UCUUCU,UCUUGC,UCUUUC,UUGUGC,UUGUGG |
RBP on BSJ (Exon and Intron)
Plot
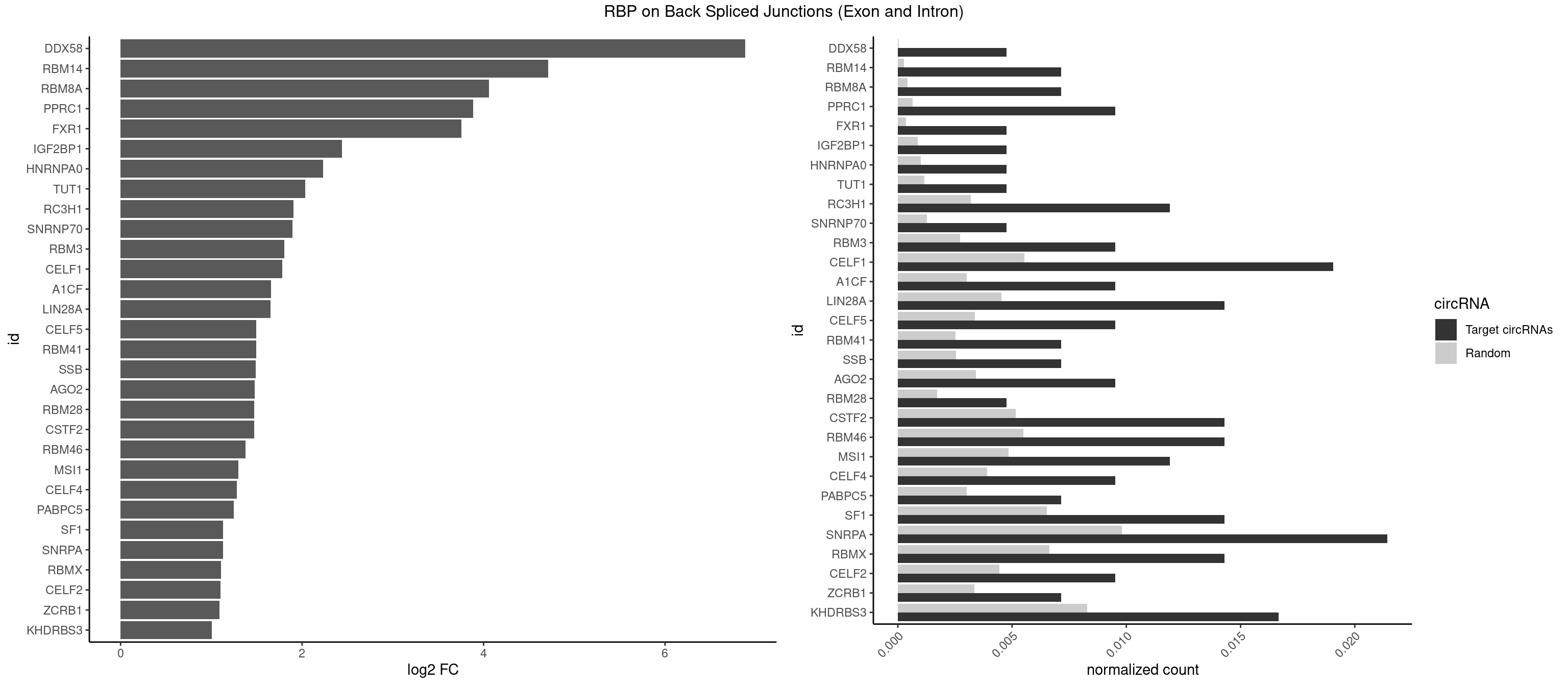
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| DDX58 | 1 | 7 | 0.004761905 | 4.030633e-05 | 6.884389 | GCGCGC | GCGCGC |
| RBM14 | 2 | 53 | 0.007142857 | 2.720677e-04 | 4.714464 | CGCGCC,GCGCGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBM8A | 2 | 84 | 0.007142857 | 4.282547e-04 | 4.059960 | CGCGCC,GCGCGC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| PPRC1 | 3 | 127 | 0.009523810 | 6.449012e-04 | 3.884389 | CGCGCC,GCGCGC,GGCGCG | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| FXR1 | 1 | 69 | 0.004761905 | 3.526804e-04 | 3.755106 | ACGACG | ACGACA,ACGACG,AUGACA,AUGACG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 8.766626e-04 | 2.441445 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPA0 | 1 | 200 | 0.004761905 | 1.012696e-03 | 2.233337 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| TUT1 | 1 | 230 | 0.004761905 | 1.163845e-03 | 2.032640 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RC3H1 | 4 | 631 | 0.011904762 | 3.184200e-03 | 1.902536 | CCUUCU,CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SNRNP70 | 1 | 253 | 0.004761905 | 1.279726e-03 | 1.895704 | AUUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBM3 | 3 | 541 | 0.009523810 | 2.730754e-03 | 1.802240 | AAAACU,AAACUA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| CELF1 | 7 | 1097 | 0.019047619 | 5.532044e-03 | 1.783726 | GUGUUG,GUUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| A1CF | 3 | 598 | 0.009523810 | 3.017936e-03 | 1.657976 | UAAUUA,UGAUCA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| LIN28A | 5 | 900 | 0.014285714 | 4.539500e-03 | 1.653968 | AGGAGA,GGAGAA,GGAGAU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| CELF5 | 3 | 669 | 0.009523810 | 3.375655e-03 | 1.496371 | GUGUUG,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM41 | 2 | 502 | 0.007142857 | 2.534260e-03 | 1.494937 | UACUUG,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| SSB | 2 | 505 | 0.007142857 | 2.549375e-03 | 1.486358 | GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| AGO2 | 3 | 677 | 0.009523810 | 3.415961e-03 | 1.479247 | AAAGUG,GUGCUU,UAAAGU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| RBM28 | 1 | 340 | 0.004761905 | 1.718057e-03 | 1.470761 | AGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| CSTF2 | 5 | 1022 | 0.014285714 | 5.154172e-03 | 1.470761 | GUGUUG,GUGUUU,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| RBM46 | 5 | 1091 | 0.014285714 | 5.501814e-03 | 1.376594 | AUCAUG,AUCAUU,AUGAAA,AUGAUG,GAUCAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| MSI1 | 4 | 961 | 0.011904762 | 4.846836e-03 | 1.296424 | AGUAAG,UAGGAG,UAGUAA,UAGUAG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| CELF4 | 3 | 776 | 0.009523810 | 3.914752e-03 | 1.282618 | GUGUUG,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC5 | 2 | 596 | 0.007142857 | 3.007860e-03 | 1.247764 | AGAAAU,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SF1 | 5 | 1294 | 0.014285714 | 6.524587e-03 | 1.130615 | AGUAAC,AGUAAG,CAGUCA,UACUGA,UAGUAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SNRPA | 8 | 1947 | 0.021428571 | 9.814591e-03 | 1.126536 | AGGAGA,AUGCUG,AUUCCU,GGAGAU,UGCACC,UUCCUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| RBMX | 5 | 1316 | 0.014285714 | 6.635429e-03 | 1.106311 | AAGUGU,ACCAAA,AGUAAC,AGUGUU,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF2 | 3 | 881 | 0.009523810 | 4.443773e-03 | 1.099754 | UAUGUG,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 3.360540e-03 | 1.087808 | AUUUAA,GCUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| KHDRBS3 | 6 | 1646 | 0.016666667 | 8.298065e-03 | 1.006119 | AAAUAA,AAUAAA,AGAUAA,AUAAAG,AUUAAA,UAAAAC | AAAUAA,AAUAAA,AGAUAA,AUAAAA,AUAAAC,AUAAAG,AUUAAA,GAUAAA,UAAAAC,UAAAUA,UUAAAC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.