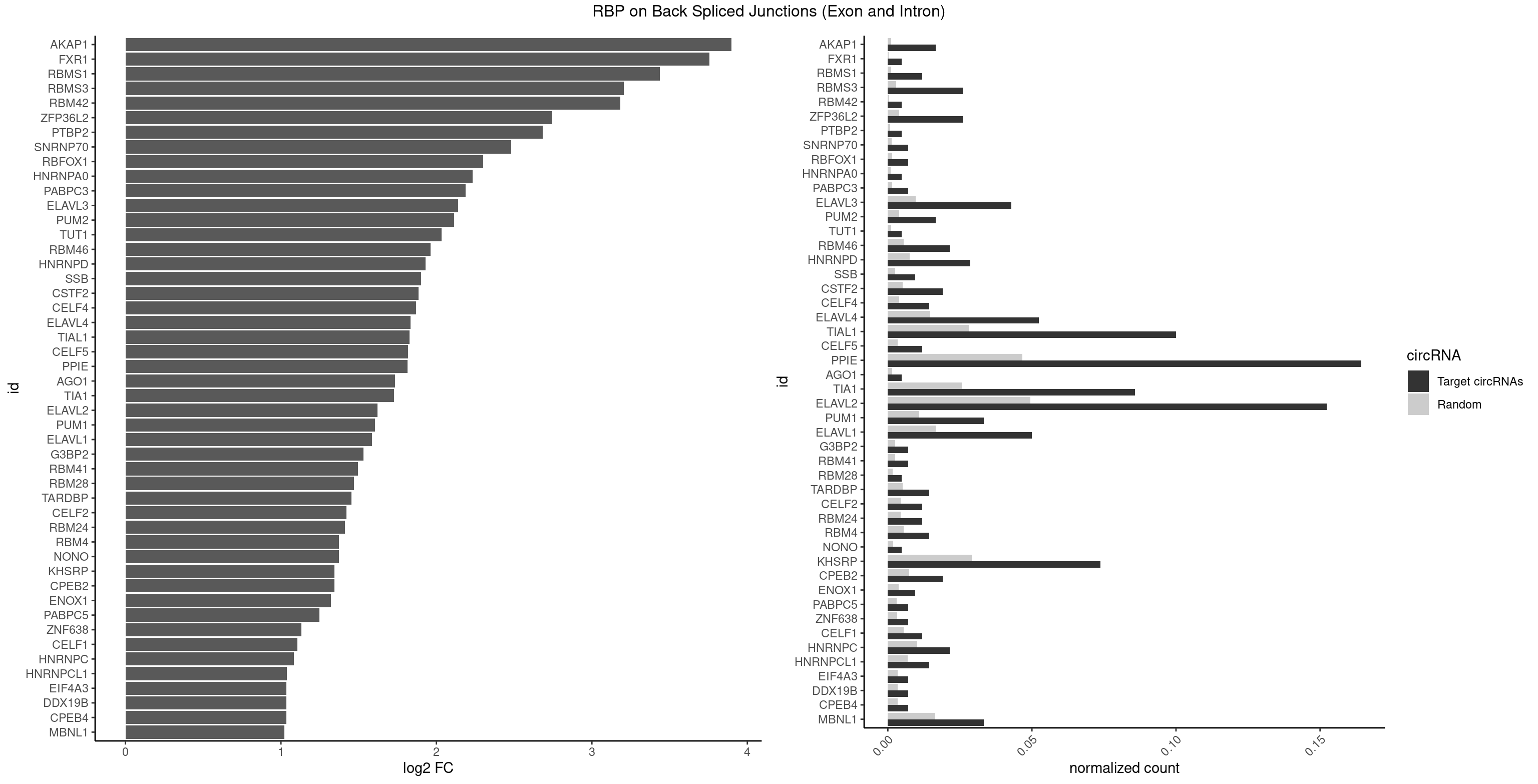
| AKAP1 |
6 |
221 |
0.016666667 |
0.0011185006 |
3.897328 |
AUAUAU,UAUAUA |
AUAUAU,UAUAUA |
| FXR1 |
1 |
69 |
0.004761905 |
0.0003526804 |
3.755106 |
AUGACA |
ACGACA,ACGACG,AUGACA,AUGACG |
| RBMS1 |
4 |
217 |
0.011904762 |
0.0010983474 |
3.438132 |
AUAUAG,UAUAUA |
AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| RBMS3 |
10 |
562 |
0.026190476 |
0.0028365578 |
3.206829 |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAU,UAUAUA |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBM42 |
1 |
103 |
0.004761905 |
0.0005239823 |
3.183949 |
AACUAA |
AACUAA,AACUAC,ACUAAG,ACUACG |
| ZFP36L2 |
10 |
774 |
0.026190476 |
0.0039046755 |
2.745768 |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| PTBP2 |
1 |
146 |
0.004761905 |
0.0007406288 |
2.684716 |
CUCUCU |
CUCUCU |
| SNRNP70 |
2 |
253 |
0.007142857 |
0.0012797259 |
2.480666 |
AUUCAA,UUCAAG |
AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBFOX1 |
2 |
287 |
0.007142857 |
0.0014510278 |
2.299426 |
AGCAUG,GCAUGA |
AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA0 |
1 |
200 |
0.004761905 |
0.0010126965 |
2.233337 |
AAUUUA |
AAUUUA,AGAUAU,AGUAGG |
| PABPC3 |
2 |
310 |
0.007142857 |
0.0015669085 |
2.188580 |
AAAACC,GAAAAC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| ELAVL3 |
17 |
1928 |
0.042857143 |
0.0097188634 |
2.140676 |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
AUUUAU,AUUUUA,UAUUUA,UAUUUU,UUAUUU,UUUAUU,UUUUUU |
| PUM2 |
6 |
764 |
0.016666667 |
0.0038542926 |
2.112428 |
UACAUA,UAUAUA,UGUAGA,UGUAUA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| TUT1 |
1 |
230 |
0.004761905 |
0.0011638452 |
2.032640 |
AAUACU |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM46 |
8 |
1091 |
0.021428571 |
0.0055018138 |
1.961556 |
AAUGAA,AAUGAU,AUGAAA,AUGAAU,AUGAUG,AUGAUU,GAUGAU |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPD |
11 |
1488 |
0.028571429 |
0.0075020153 |
1.929223 |
AAUUUA,AUUUAU,UAUUUA,UUAUUU,UUUAUU |
AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| SSB |
3 |
505 |
0.009523810 |
0.0025493753 |
1.901395 |
CUGUUU,UGUUUU |
CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| CSTF2 |
7 |
1022 |
0.019047619 |
0.0051541717 |
1.885798 |
GUGUGU,GUUUUG,UGUGUG,UGUUUU |
GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF4 |
5 |
776 |
0.014285714 |
0.0039147521 |
1.867580 |
GGUGUG,GUGUGU,UGUGUG |
GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ELAVL4 |
21 |
2916 |
0.052380952 |
0.0146966949 |
1.833551 |
AUUUAU,UAUUUA,UAUUUU,UUAUUU,UUGUAU,UUUAUU,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
| TIAL1 |
41 |
5597 |
0.100000000 |
0.0282043531 |
1.826010 |
AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUU,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUG,UUUUUU |
AAAUUU,AAUUUU,AGUUUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAGUUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUC,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUAUUU,UUCAGU,UUUAAA,UUUAUU,UUUCAG,UUUUAA,UUUUAU,UUUUCA,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| CELF5 |
4 |
669 |
0.011904762 |
0.0033756550 |
1.818299 |
GUGUGU,UGUGUG |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PPIE |
68 |
9262 |
0.164285714 |
0.0466696896 |
1.815649 |
AAAAUA,AAAUAU,AAAUUA,AAUAAU,AAUAUA,AAUAUU,AAUUAU,AAUUUA,AAUUUU,AUAAAU,AUAAUA,AUAUAA,AUAUAU,AUAUUU,AUUAUA,AUUUAU,AUUUUA,AUUUUU,UAAAAU,UAAAUU,UAAUAU,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUUA,UAUUUU,UUAAAA,UUAAUA,UUAUAA,UUAUAU,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| AGO1 |
1 |
283 |
0.004761905 |
0.0014308746 |
1.734641 |
AGGUAG |
AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| TIA1 |
35 |
5140 |
0.085714286 |
0.0259018541 |
1.726480 |
AUUUUC,AUUUUG,AUUUUU,CUCCUU,CUUUUA,CUUUUC,CUUUUU,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCU,UUUUGU,UUUUUA,UUUUUG,UUUUUU |
AUUUUC,AUUUUG,AUUUUU,CCUUUC,CUCCUU,CUUUUA,CUUUUC,CUUUUG,CUUUUU,GUUUUA,GUUUUC,GUUUUG,GUUUUU,UAUUUU,UCCUUU,UCUCCU,UUAUUU,UUCUCC,UUUAUU,UUUUAU,UUUUCG,UUUUCU,UUUUGG,UUUUGU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| ELAVL2 |
63 |
9826 |
0.152380952 |
0.0495112858 |
1.621853 |
AAUUUA,AAUUUU,AUACUU,AUAUUU,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUUA,CUUUUU,GAUUUU,GUUUUU,UAAUUU,UAUAUA,UAUAUU,UAUUUA,UAUUUG,UAUUUU,UCUUUU,UGAUUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCUUU,UUGUAU,UUUAAG,UUUAAU,UUUAUA,UUUAUU,UUUCUU,UUUUAA,UUUUAU,UUUUUA,UUUUUG,UUUUUU |
AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| PUM1 |
13 |
2172 |
0.033333333 |
0.0109482064 |
1.606271 |
AAUAUU,AAUUGU,AUUGUA,UAAUAU,UACAUA,UAUAUA,UGUAGA,UGUAUA,UUUAAU |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| ELAVL1 |
20 |
3309 |
0.050000000 |
0.0166767432 |
1.584091 |
AUUUAU,UAUUUA,UAUUUU,UGAUUU,UGUUUU,UUAUUU,UUGUUU,UUUAUU,UUUGUU,UUUUUG,UUUUUU |
AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| G3BP2 |
2 |
490 |
0.007142857 |
0.0024738009 |
1.529772 |
AGGAUG,GGAUGA |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM41 |
2 |
502 |
0.007142857 |
0.0025342604 |
1.494937 |
AUACUU,UUACAU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| RBM28 |
1 |
340 |
0.004761905 |
0.0017180572 |
1.470761 |
UGUAGA |
AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| TARDBP |
5 |
1034 |
0.014285714 |
0.0052146312 |
1.453936 |
GUGUGU,GUUUUG,UGUGUG |
GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CELF2 |
4 |
881 |
0.011904762 |
0.0044437727 |
1.421682 |
GUGUGU,UGUGUG |
AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| RBM24 |
4 |
888 |
0.011904762 |
0.0044790407 |
1.410277 |
GUGUGU,UGUGUG |
AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| NONO |
1 |
364 |
0.004761905 |
0.0018389762 |
1.372636 |
AGAGGA |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM4 |
5 |
1094 |
0.014285714 |
0.0055169287 |
1.372636 |
CCUUCC,CUUCCU,CUUCUU,UCCUUC,UUCUUG |
CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| KHSRP |
30 |
5764 |
0.073809524 |
0.0290457477 |
1.345480 |
AUAUUU,AUUUAU,AUUUUA,CCUUCC,CUCCUU,CUGCCU,GUGUGU,UAUAUU,UAUUUA,UAUUUU,UCCUUC,UGCCUC,UGUAUA,UGUGUG,UUAUUU,UUGUAU,UUUAUA,UUUAUU |
ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| CPEB2 |
7 |
1487 |
0.019047619 |
0.0074969770 |
1.345230 |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
AUUUUU,CAUUUU,CCUUUU,CUUUUU,UUUUUU |
| ENOX1 |
3 |
756 |
0.009523810 |
0.0038139863 |
1.320239 |
AAUACA,AUACAG,UAUACA |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PABPC5 |
2 |
596 |
0.007142857 |
0.0030078597 |
1.247764 |
AGAAAA,GAAAAU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| ZNF638 |
2 |
646 |
0.007142857 |
0.0032597743 |
1.131729 |
GGUUGG,GUUGGU |
CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF1 |
4 |
1097 |
0.011904762 |
0.0055320435 |
1.105654 |
GUGUGU,UGUGUG |
CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPC |
8 |
2006 |
0.021428571 |
0.0101118501 |
1.083489 |
AUUUUU,CUUUUU,UUUUUA,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| HNRNPCL1 |
5 |
1381 |
0.014285714 |
0.0069629182 |
1.036809 |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
AUUUUU,CUUUUU,UUUUUG,UUUUUU |
| CPEB4 |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |
| DDX19B |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |
| EIF4A3 |
2 |
691 |
0.007142857 |
0.0034864974 |
1.034723 |
UUUUUU |
UUUUUU |
| MBNL1 |
13 |
3258 |
0.033333333 |
0.0164197904 |
1.021530 |
AUGCCA,AUGCUU,CUGCCU,CUUGCU,GCUUUU,GUGCUC,UCUCGC,UGCUUU,UUGCUG,UUGCUU,UUUGCU |
ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |