circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000124788:-:6:16326393:16328470
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000124788:-:6:16326393:16328470 | ENSG00000124788 | ENST00000244769 | - | 6 | 16326394 | 16328470 | 2077 | CAUCCAGAGCUGCUGUUGGCGGAUUGUACCCACGGGGAGAUGAUUCCUCAUGAAGAGCCUGGAUCCCCUACAGAAAUCAAAUGUGACUUUCCGUUUAUCAGACUAAAAUCAGAGCCAUCCAGACAGUGAAACAGUCACCGUGGAGGGGGGACGGCGAAAAAUGAAAUCCAACCAAGAGCGGAGCAACGAAUGCCUGCCUCCCAAGAAGCGCGAGAUCCCCGCCACCAGCCGGUCCUCCGAGGAGAAGGCCCCUACCCUGCCCAGCGACAACCACCGGGUGGAGGGCACAGCAUGGCUCCCGGGCAACCCUGGUGGCCGGGGCCACGGGGGCGGGAGGCAUGGGCCGGCAGGGACCUCGGUGGAGCUUGGUUUACAACAGGGAAUAGGUUUACACAAAGCAUUGUCCACAGGGCUGGACUACUCCCCGCCCAGCGCUCCCAGGUCUGUCCCCGUGGCCACCACGCUGCCUGCCGCGUACGCCACCCCGCAGCCAGGGACCCCGGUGUCCCCCGUGCAGUACGCUCACCUGCCGCACACCUUCCAGUUCAUUGGGUCCUCCCAAUACAGUGGAACCUAUGCCAGCUUCAUCCCAUCACAGCUGAUCCCCCCAACCGCCAACCCCGUCACCAGUGCAGUGGCCUCGGCCGCAGGGGCCACCACUCCAUCCCAGCGCUCCCAGCUGGAGGCCUAUUCCACUCUGCUGGCCAACAUGGGCAGUCUGAGCCAGACGCCGGGACACAAGGCUGAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAUCAGCAUCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCAGCACCUCAGCAGGGCUCCGGGGCUCAUCACCCCGGGGUCCCCCCCACCAGCCCAGCAGAACCAGUACGUCCACAUUUCCAGUUCUCCGCAGAACACCGGCCGCACCGCCUCUCCUCCGGCCAUCCCCGUCCACCUCCACCCCCACCAGACGAUGAUCCCACACACGCUCACCCUGGGGCCCCCCUCCCAGGUCGUCAUGCAAUACGCCGACUCCGGCAGCCACUUUGUCCCUCGGGAGGCCACCAAGAAAGCUGAGAGCAGCCGGCUGCAGCAGGCCAUCCAGGCCAAGGAGGUCCUGAACGGUGAGAUGGAGAAGAGCCGGCGGUACGGGGCCCCGUCCUCAGCCGACCUGGGCCUGGGCAAGGCAGGCGGCAAGUCGGUUCCUCACCCGUACGAGUCCAGGCACGUGGUGGUCCACCCGAGCCCCUCAGACUACAGCAGUCGUGAUCCUUCGGGGGUCCGGGCCUCUGUGAUGGUCCUGCCCAACAGCAACACGCCCGCAGCUGACCUGGAGGUGCAACAGGCCACUCAUCGUGAAGCCUCCCCUUCUACCCUCAACGACAAAAGUGGCCUGCAUUUAGGGAAGCCUGGCCACCGGUCCUACGCGCUCUCACCCCACACGGUCAUUCAGACCACACACAGUGCUUCAGAGCCACUCCCGGUGGGACUGCCAGCCACGGCCUUCUACGCAGGGACUCAACCCCCUGUCAUCGGCUACCUGAGCGGCCAGCAGCAAGCAAUCACCUACGCCGGCAGCCUGCCCCAGCACCUGGUGAUCCCCGGCACACAGCCCCUGCUCAUCCCGGUCGGCAGCACUGACAUGGAAGCGUCGGGGGCAGCCCCGGCCAUAGUCACGUCAUCCCCCCAGUUUGCUGCAGUGCCUCACACGUUCGUCACCACCGCCCUUCCCAAGAGCGAGAACUUCAACCCUGAGGCCCUGGUCACCCAGGCCGCCUACCCAGCCAUGGUGCAGGCCCAGAUCCACCUGCCUGUGGUGCAGUCCGUGGCCUCCCCGGCGGCGGCUCCCCCUACGCUGCCUCCCUACUUCAUGAAAGGCUCCAUCAUCCAGUUGGCCAACGGGGAGCUAAAGAAGGUGGAAGACUUAAAAACAGAAGAUUUCAUCCAGAGUGCAGAGAUAAGCAACGACCUGAAGAUCGACUCCAGCACCGUAGAGAGGAUUGAAGACAGCCAUAGCCCGGGCGUGGCCGUGAUACAGUUCGCCGUCGGGGAGCACCGAGCCCAGCAUCCAGAGCUGCUGUUGGCGGAUUGUACCCACGGGGAGAUGAUUCCUCA | circ |
| ENSG00000124788:-:6:16326393:16328470 | ENSG00000124788 | ENST00000244769 | - | 6 | 16326394 | 16328470 | 22 | ACCGAGCCCAGCAUCCAGAGCU | bsj |
| ENSG00000124788:-:6:16326393:16328470 | ENSG00000124788 | ENST00000244769 | - | 6 | 16328461 | 16328670 | 210 | CCAAAGUGUUGGCUAGUGUUUUCUCUGCUUCAGUGCUUGGGGUAUGAUUGGGUUAUGGGAGUUCACACCGAGUCCAGGGCCUAGUCUUAAUCUUGCCAAAGAUGUUCUUUCCCCGGUGCUCAUGUUCUGAUGUCCUUUCCCUCCUUCCCUUUCUCCUCCCUUUCCUUUUCCCUUUGUCACUGCCCUCUUCCCUUUCCCAGCAUCCAGAGC | ie_up |
| ENSG00000124788:-:6:16326393:16328470 | ENSG00000124788 | ENST00000244769 | - | 6 | 16326194 | 16326403 | 210 | CCGAGCCCAGGUAACGUUAGCCAGGGUGGCACAGGGAUGGGACACCACACCGUGAUGCCAUCAUCAUCUCCUGGCAAGACGAAUUGCUUCUAUGAGGCAGGAUUAAGGGUUCUCGGGUACACCUAGACCUUAGACUCGGCCUUUCCCAACUGCGUUCUCUAGAAAAAAUAAGCCCCAUUUCCCCGUGAUCUCUGCUGUGUGUAAUGAAUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
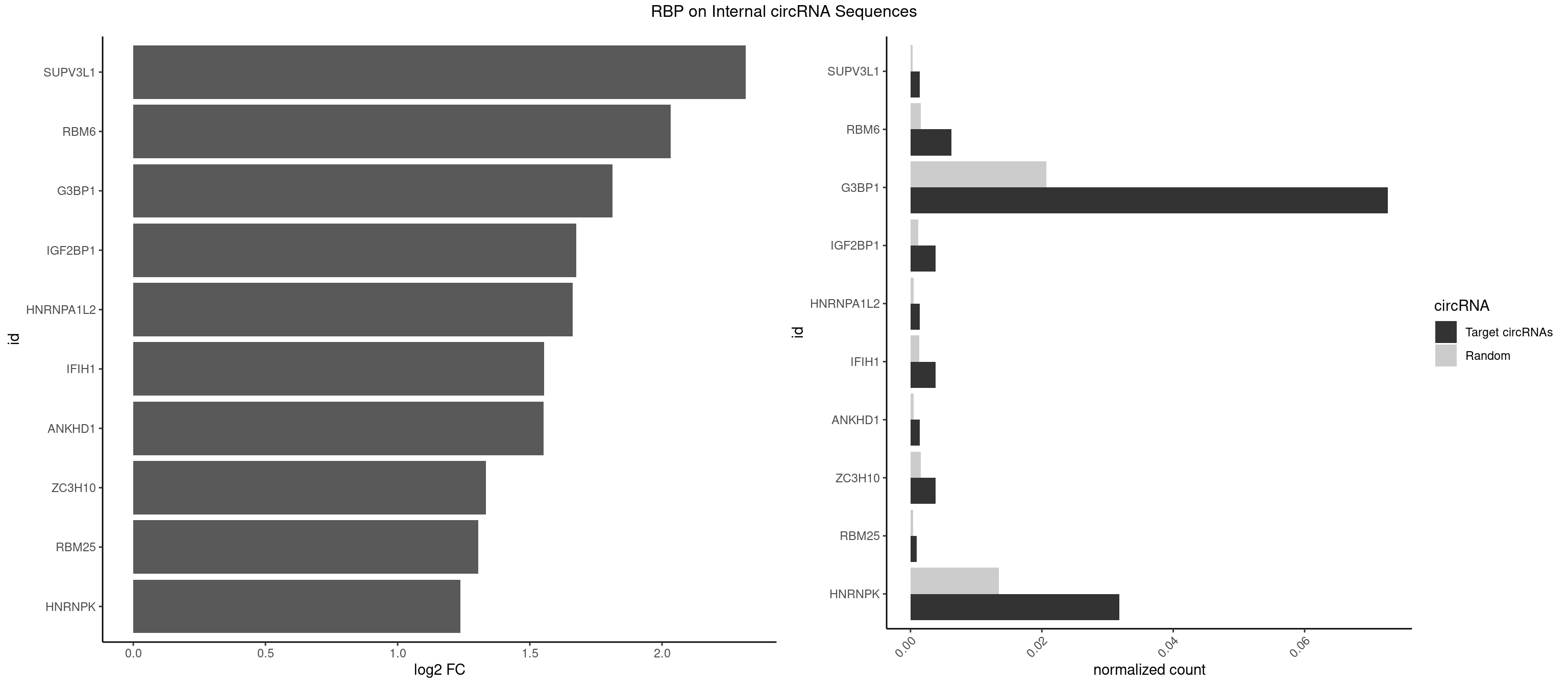
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 2 | 199 | 0.0014443909 | 0.0002898408 | 2.317129 | CCGCCC | CCGCCC |
| RBM6 | 12 | 1054 | 0.0062590274 | 0.0015289102 | 2.033435 | AAUCCA,AUCCAA,AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| G3BP1 | 150 | 14282 | 0.0727010111 | 0.0206989801 | 1.812416 | ACACGC,ACAGGC,ACCCAC,ACCCCC,ACCGGC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCC,CAGGCA,CAGGCC,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAG,CCAUCC,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGGCA,CCGGCC,CCUACG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCG,CUACGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UCCGCA,UCGGCA,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| IGF2BP1 | 7 | 831 | 0.0038517092 | 0.0012057377 | 1.675583 | ACCCGU,AGCACC,CACCCG | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPA1L2 | 2 | 314 | 0.0014443909 | 0.0004564992 | 1.661777 | UAGGGA,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| IFIH1 | 7 | 904 | 0.0038517092 | 0.0013115296 | 1.554248 | GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ANKHD1 | 2 | 339 | 0.0014443909 | 0.0004927293 | 1.551594 | AGACGA,GACGAU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| ZC3H10 | 7 | 1053 | 0.0038517092 | 0.0015274610 | 1.334363 | CAGCGA,CAGCGC,CCAGCG,GAGCGA | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| RBM25 | 1 | 268 | 0.0009629273 | 0.0003898359 | 1.304560 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| HNRNPK | 65 | 9304 | 0.0317766009 | 0.0134848428 | 1.236626 | ACCCCA,ACCCCC,ACCCGA,ACCCGU,CAACCC,CAUCCC,CCAACC,CCAUCC,CCCCAA,CCCCAC,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCC,UCCCCU,UCCCUA | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
RBP on BSJ (Exon Only)
Plot
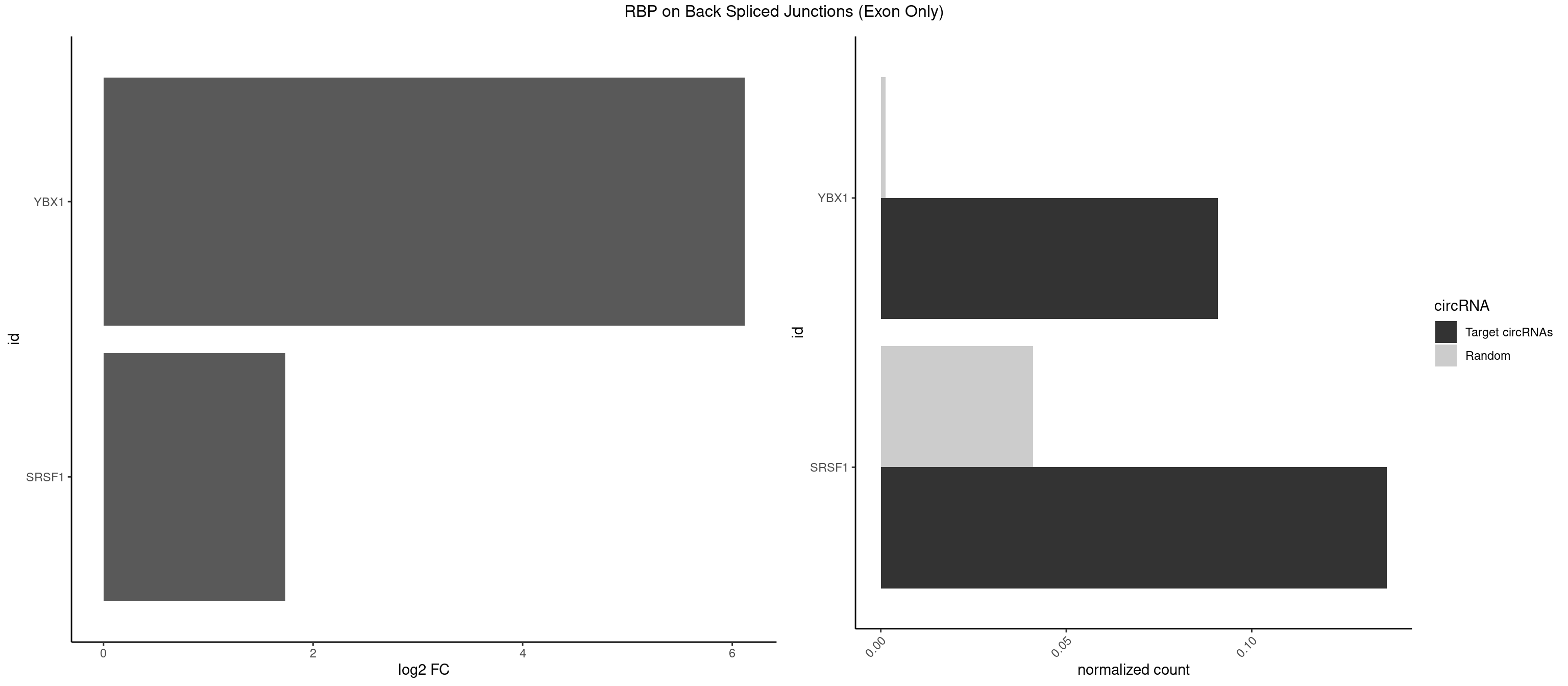
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| YBX1 | 1 | 19 | 0.09090909 | 0.001309929 | 6.116864 | CCAGCA | AACCAC,ACACCA,ACAUCU,ACCACC,CACCAC,CAGCAA,CCACCA,CCAGCA,CCUGCG,CUGCGG,GAUCUG,GCCUGC,UCCAGC,UGCGGU |
| SRSF1 | 2 | 625 | 0.13636364 | 0.041000786 | 1.733736 | CCAGCA,CCCAGC | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
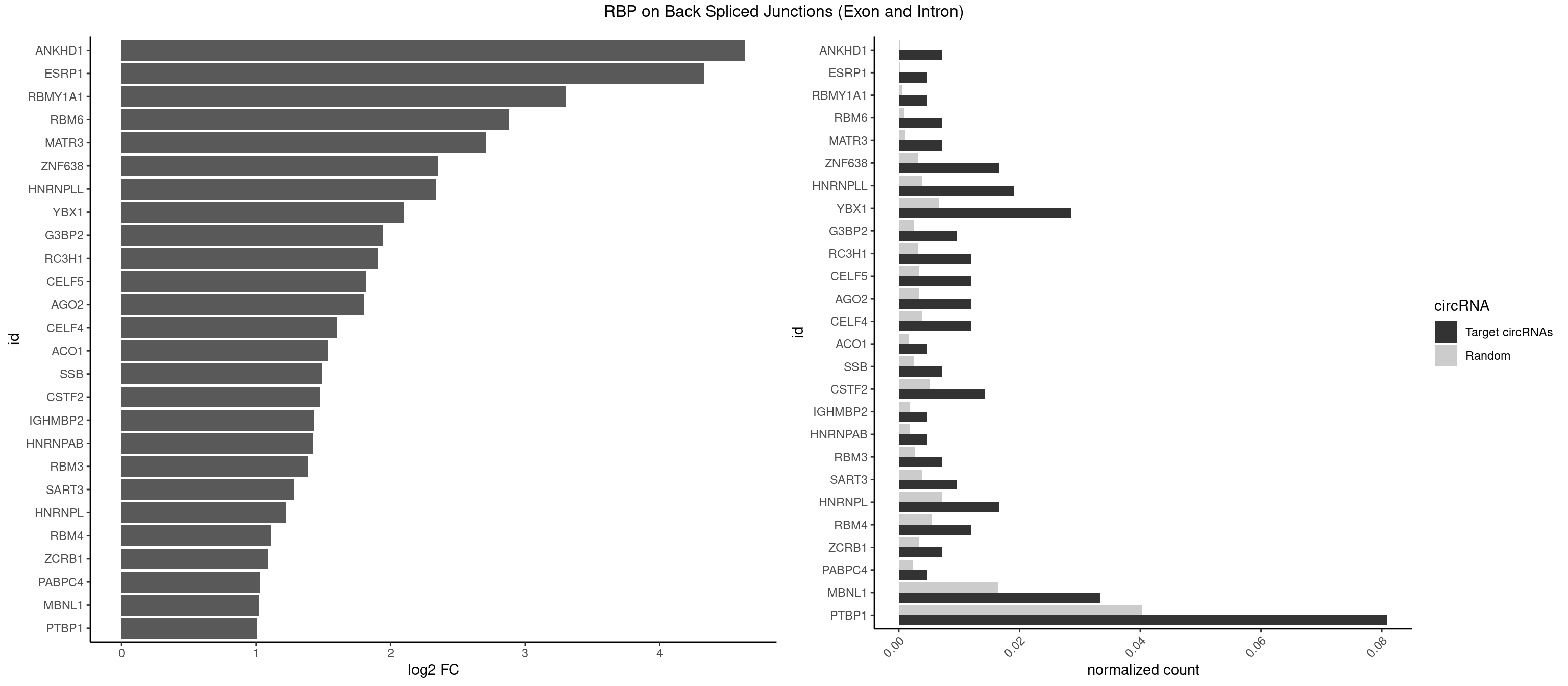
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ANKHD1 | 2 | 56 | 0.007142857 | 0.0002871826 | 4.636461 | AGACGA,GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| ESRP1 | 1 | 46 | 0.004761905 | 0.0002367997 | 4.329800 | AGGGAU | AGGGAU |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | CAAGAC | ACAAGA,CAAGAC |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ZNF638 | 6 | 646 | 0.016666667 | 0.0032597743 | 2.354122 | CGUUCU,GGUUCU,GUUCUU,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| HNRNPLL | 7 | 748 | 0.019047619 | 0.0037736800 | 2.335567 | ACACCA,ACCACA,AGACGA,CACACC,CACCAC,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| YBX1 | 11 | 1321 | 0.028571429 | 0.0066606207 | 2.100845 | ACACCA,ACCACA,AUCAUC,CACACC,CACCAC,CAUCAU,CCACAC,CCAGCA | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUU,GGAUGG,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RC3H1 | 4 | 631 | 0.011904762 | 0.0031841999 | 1.902536 | UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| CELF5 | 4 | 669 | 0.011904762 | 0.0033756550 | 1.818299 | GUGUGU,GUGUUG,GUGUUU,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| AGO2 | 4 | 677 | 0.011904762 | 0.0034159613 | 1.801175 | AAAAAA,AAAGUG,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GUGUGU,GUGUUG,GUGUUU,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| CSTF2 | 5 | 1022 | 0.014285714 | 0.0051541717 | 1.470761 | GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAGACG,AGACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AAAAAA,AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| HNRNPL | 6 | 1419 | 0.016666667 | 0.0071543732 | 1.220068 | AAAUAA,ACACCA,ACCACA,CACACC,CACCAC | AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUCUU,CCUUCC,UCCUUC,UUCCUU | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PABPC4 | 1 | 462 | 0.004761905 | 0.0023327287 | 1.029520 | AAAAAA | AAAAAA,AAAAAG |
| MBNL1 | 13 | 3258 | 0.033333333 | 0.0164197904 | 1.021530 | AUGCCA,CUGCCC,CUGCUG,CUGCUU,GUGCUC,GUGCUU,UCUGCU,UGCUGU,UGCUUC,UUGCCA,UUGCUU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| PTBP1 | 33 | 8003 | 0.080952381 | 0.0403264813 | 1.005346 | CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUCUUC,CUUUCC,CUUUCU,CUUUUC,GCUGUG,GUCUUA,UCUUUC,UUCCCC,UUCCUU,UUCUCU,UUCUUU,UUUCCC,UUUUCC,UUUUCU | ACUUUC,ACUUUU,AGCUGU,AUCUUC,AUUUUC,AUUUUU,CAUCUU,CCAUCU,CCCCCC,CCUCUU,CCUUCC,CCUUUC,CCUUUU,CUAUCU,CUCCAU,CUCCCC,CUCUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUCCU,CUUCUC,CUUCUU,CUUUCC,CUUUCU,CUUUUC,CUUUUU,GCUCCC,GCUGUG,GGCUCC,GGGGGG,GUCUUA,GUCUUU,UACUUU,UAGCUG,UAUUUU,UCCAUC,UCCCCC,UCCUCU,UCUAUC,UCUCUC,UCUCUU,UCUUCU,UCUUUC,UCUUUU,UUACUU,UUAUUU,UUCCCC,UUCCUU,UUCUCU,UUCUUC,UUCUUG,UUCUUU,UUUCCC,UUUCUU,UUUUCC,UUUUCU,UUUUUC,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.