circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000141337:+:17:68306942:68401450
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000141337:+:17:68306942:68401450 | ENSG00000141337 | ENST00000448504 | + | 17 | 68306943 | 68401450 | 1854 | GAAACCUUACAGAAACAUGAAGCCCUCAACCAUCUGCUACUCAGUUAUUCGGGGCUGACGGCGGCUUCUAGAACAUCCAGGUGUUCUGCAGAUGCGAGAACUCAUCCUGUAGUCACCAGAUGGAGUCCCAAACAGCCAAGCAGAUGUAAGGCCUGUGCUGUGGCUCUGAGGCCCUGAAUACAGAAGGGUCACUUUCUUAGUGGCCAAAGAGCAGUUGUUGACAUUGAUGUCUAAUUAUUGAACACGACCAGUCAUUUUACUGAGCUGCGGUGAGGAAACACUGACCAUAGAAGAUCAAGCCAAAUGAGGGAUUGCAAAUUUCCUGAUUCUUUUGAAUUAGGAUUCCAGAUGGGGGCCUCAUUUCUACAGCCCCCAACAUUCCUAUAGCCGUUAUCACUGCCAUCACCACUGCCACCAGCAUCUUCUUGCAGAUUCCACCCCUGCUCCCCAGAGACUUCCUGCUUUGAAAGUGAGCAGAAAGGAAGCUCUCAGAAAAAUCUCUAGUGGUGGCUGCCGUCGCUCCAGACAAUCGGAAUCCUGCCUUCACCACCAUGGGCUGGCUUUUUCUAAAGGUUUUGUUGGCGGGAGUGAGUUUCUCAGGAUUUCUUUAUCCUCUUGUGGAUUUUUGCAUCAGUGGGAAAACAAGAGGACAGAAGCCAAACUUUGUGAUUAUUUUGGCCGAUGACAUGGGGUGGGGUGACCUGGGAGCAAACUGGGCAGAAACAAAGGACACUGCCAACCUUGAUAAGAUGGCUUCGGAGGGAAUGAGGUUUGUGGAUUUCCAUGCAGCUGCCUCCACCUGCUCACCCUCCCGGGCUUCCUUGCUCACCGGCCGGCUUGGCCUUCGCAAUGGAGUCACACGCAACUUUGCAGUCACUUCUGUGGGAGGCCUUCCGCUCAACGAGACCACCUUGGCAGAGGUGCUGCAGCAGGCGGGUUACGUCACUGGGAUAAUAGGCAAAUGGCAUCUUGGACACCACGGCUCUUAUCACCCCAACUUCCGUGGUUUUGAUUACUACUUUGGAAUCCCAUAUAGCCAUGAUAUGGGCUGUACUGAUACUCCAGGCUACAACCACCCUCCUUGUCCAGCGUGUCCACAGGGUGAUGGACCAUCAAGGAACCUUCAAAGAGACUGUUACACUGACGUGGCCCUCCCUCUUUAUGAAAACCUCAACAUUGUGGAGCAGCCGGUGAACUUGAGCAGCCUUGCCCAGAAGUAUGCUGAGAAAGCAACCCAGUUCAUCCAGCGUGCAAGCACCAGCGGGAGGCCCUUCCUGCUCUAUGUGGCUCUGGCCCACAUGCACGUGCCCUUACCUGUGACUCAGCUACCAGCAGCGCCACGGGGCAGAAGCCUGUAUGGUGCAGGGCUCUGGGAGAUGGACAGUCUGGUGGGCCAGAUCAAGGACAAAGUUGACCACACAGUGAAGGAAAACACAUUCCUCUGGUUUACAGGAGACAAUGGCCCGUGGGCUCAGAAGUGUGAGCUAGCGGGCAGUGUGGGUCCCUUCACUGGAUUUUGGCAAACUCGUCAAGGGGGAAGUCCAGCCAAGCAGACGACCUGGGAAGGAGGGCACCGGGUCCCAGCACUGGCUUACUGGCCUGGCAGAGUUCCAGUUAAUGUCACCAGCACUGCCUUGUUAAGCGUGCUGGACAUUUUUCCAACUGUGGUAGCCCUGGCCCAGGCCAGCUUACCUCAAGGACGGCGCUUUGAUGGUGUGGACGUCUCCGAGGUGCUCUUUGGCCGGUCACAGCCUGGGCACAGGGUGCUGUUCCACCCCAACAGCGGGGCAGCUGGAGAGUUUGGAGCCCUGCAGACUGUCCGCCUGGAGCGUUACAAGGCCUUCUACAUUACCGGAAACCUUACAGAAACAUGAAGCCCUCAACCAUCUGCUACUCAGUUAUUC | circ |
| ENSG00000141337:+:17:68306942:68401450 | ENSG00000141337 | ENST00000448504 | + | 17 | 68306943 | 68401450 | 22 | CUACAUUACCGGAAACCUUACA | bsj |
| ENSG00000141337:+:17:68306942:68401450 | ENSG00000141337 | ENST00000448504 | + | 17 | 68306743 | 68306952 | 210 | UUCCAACCCUGUCGCUAUAGGUCCAUUGCGAAUAUUAUUAUCAUCACUAUUUUUCAGGUGGAGAAACUGAUGAACCAGUUAAAUAUUUGAAUAAAAACAUCUGUUGGCAUGGAAAAGUGAUUAUUUCCUAAGCUACUGAGUAAAACACAGCUGCACUGGCAGUUUUAACUCUGUUUUCUUGUCAAUUUUUGUUCUUUCAGGAAACCUUAC | ie_up |
| ENSG00000141337:+:17:68306942:68401450 | ENSG00000141337 | ENST00000448504 | + | 17 | 68401441 | 68401650 | 210 | UACAUUACCGGUGAGUGAGGGGCCACUUAGCCCUGCCUCCCACAGUCACAGCUGCACGGGGACCCCAUGGACUCUCACCCAGGCCUCUGGGAAUGAGUGUGAGUCCCUCCUUUGUGGGGGGUUCUCAGCACCUCUUGACAAGCGUCUCCUCCAAACCCAGGGUCCCUGCACAGGCUCUCUGGAUCCCUCAUAUGCAAAUAAGAUCUUUGU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
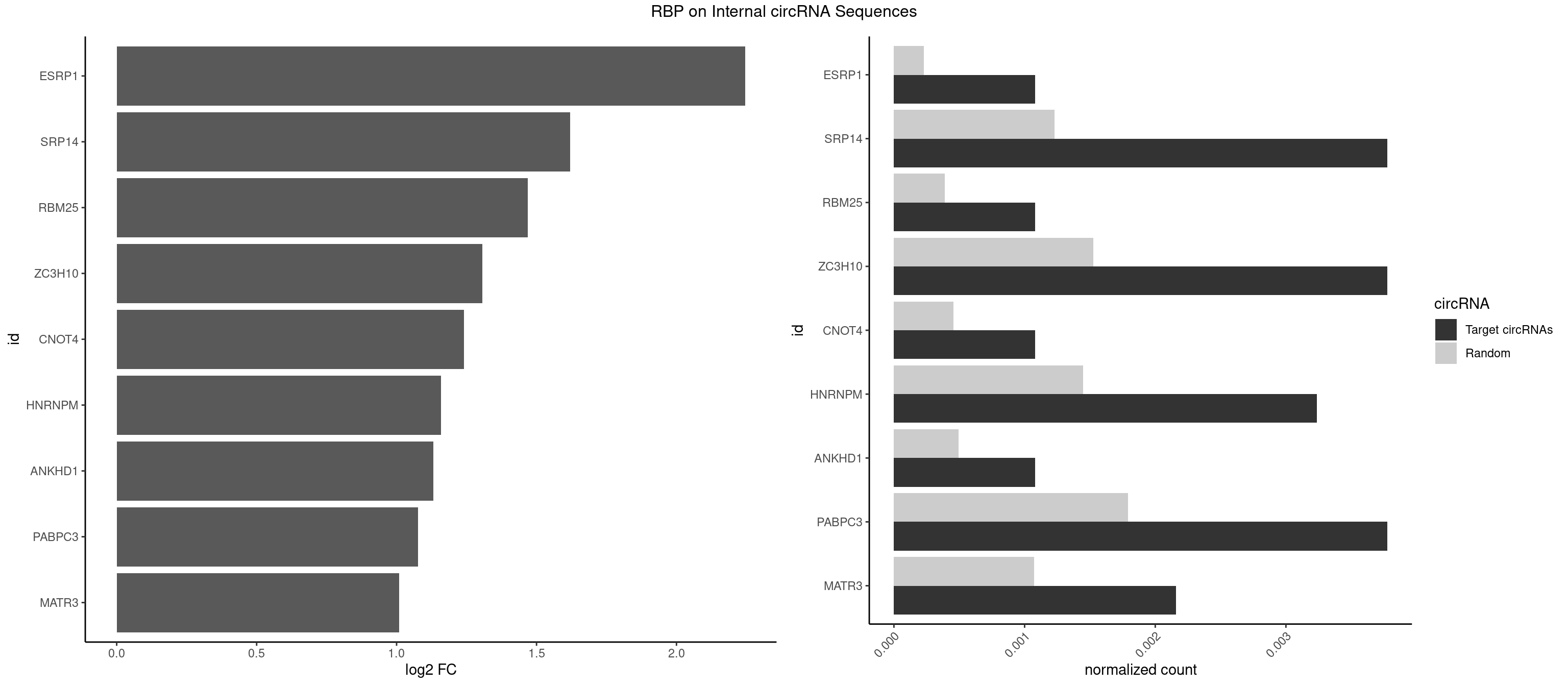
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.001078749 | 0.0002275250 | 2.245262 | AGGGAU | AGGGAU |
| SRP14 | 6 | 847 | 0.003775620 | 0.0012289250 | 1.619317 | CCUGUA,CGCCUG,CUGUAG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM25 | 1 | 268 | 0.001078749 | 0.0003898359 | 1.468420 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| ZC3H10 | 6 | 1053 | 0.003775620 | 0.0015274610 | 1.305578 | CAGCGC,CCAGCG,GCAGCG,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| CNOT4 | 1 | 314 | 0.001078749 | 0.0004564992 | 1.240674 | GACAGA | GACAGA |
| HNRNPM | 5 | 999 | 0.003236246 | 0.0014492040 | 1.159061 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ANKHD1 | 1 | 339 | 0.001078749 | 0.0004927293 | 1.130491 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| PABPC3 | 6 | 1234 | 0.003775620 | 0.0017897669 | 1.076942 | AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| MATR3 | 3 | 739 | 0.002157497 | 0.0010724109 | 1.008501 | AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
RBP on BSJ (Exon Only)
Plot
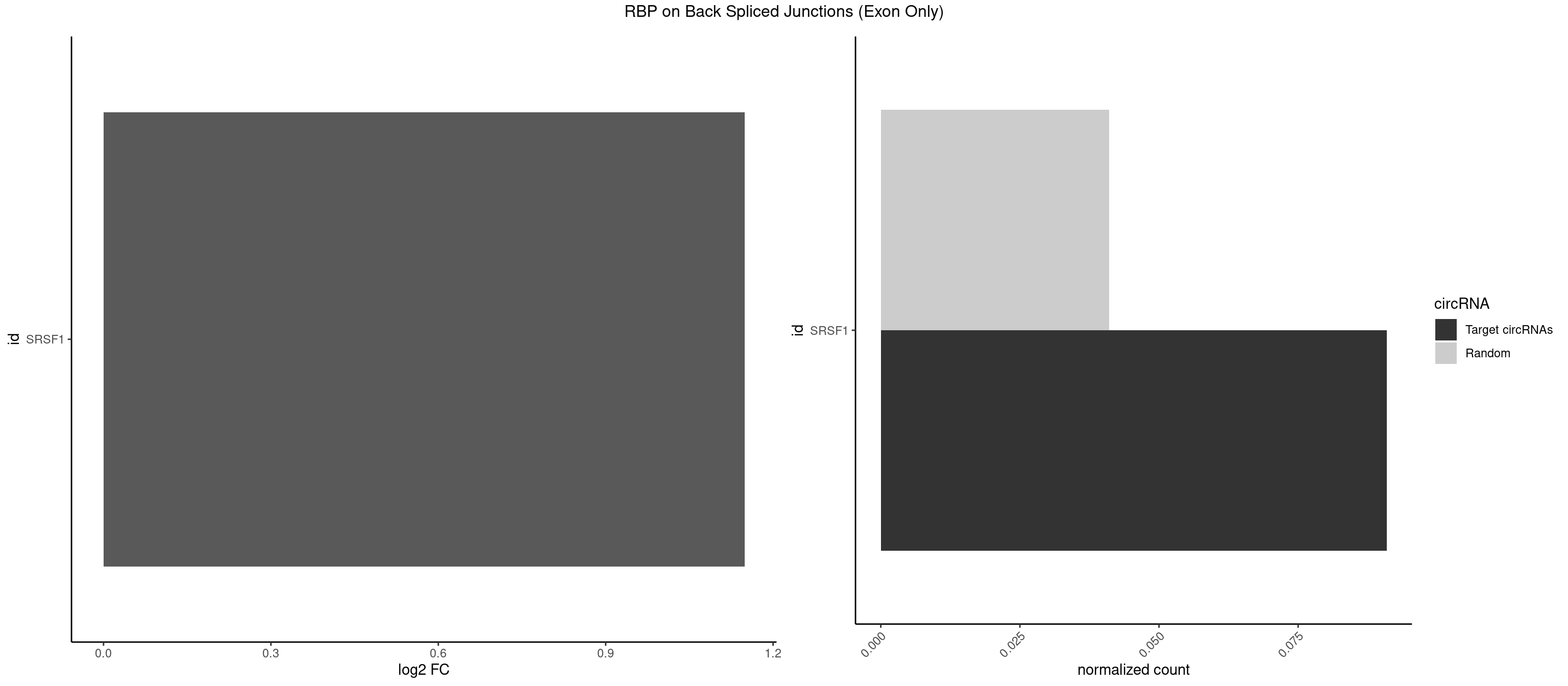
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF1 | 1 | 625 | 0.09090909 | 0.04100079 | 1.148773 | ACCGGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
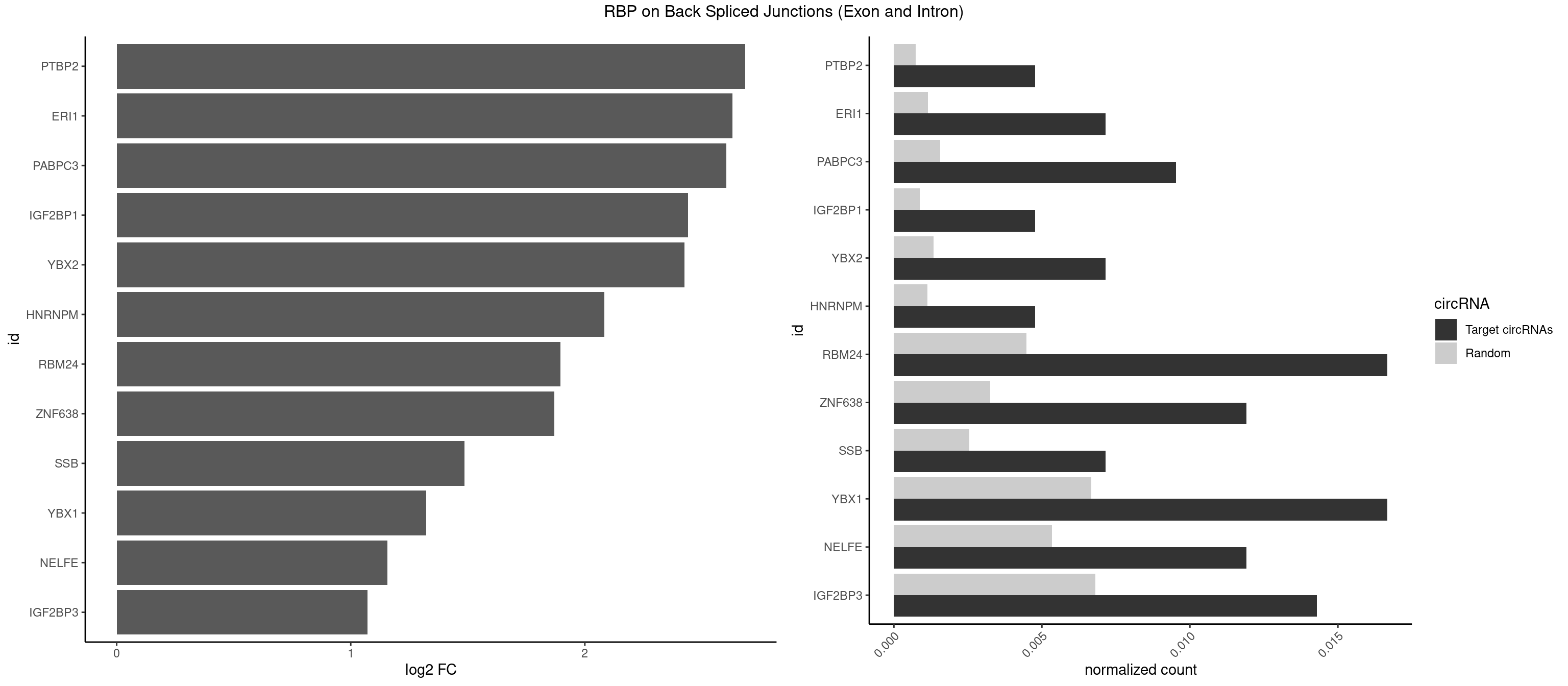
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUUCAG | UUCAGA,UUUCAG |
| PABPC3 | 3 | 310 | 0.009523810 | 0.0015669085 | 2.603618 | AAAAAC,AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | AACAUC,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| RBM24 | 6 | 888 | 0.016666667 | 0.0044790407 | 1.895704 | AGUGUG,GAGUGA,GAGUGU,GUGUGA,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ZNF638 | 4 | 646 | 0.011904762 | 0.0032597743 | 1.868695 | GGUUCU,GUUCUU,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| YBX1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | AACAUC,ACAUCU,AUCAUC,CAUCUG,CCCUGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUCUCU,CUCUGG,UCUCUG | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| IGF2BP3 | 5 | 1348 | 0.014285714 | 0.0067966546 | 1.071676 | AAAAAC,AAAACA,AAACAC,AACACA | AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.