circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000131697:-:1:5961793:5986327
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000131697:-:1:5961793:5986327 | ENSG00000131697 | ENST00000489180 | - | 1 | 5961794 | 5986327 | 711 | AUCAUCCGGGAAGCCCACGGGACCCUCAGGCGGGCAGGAUGAACGACUGGCACAGGAUCUUCACCCAAAACGUGCUUGUCCCUCCCCACCCACAGAGAGCGCGCCAGCCUUGGAAGGAAUCCACGGCAUUCCAGUGUGUCCUCAAGUGGCUGGACGGACCGGUAAUUAGGCAGGGCGUGCUGGAGGUACUGUCAGAGGUUGAAUGCCAUCUGCGAGUGUCUUUCUUUGAUGUCACCUACCGGCACUUCUUUGGGAGGACGUGGAAAACCACAGUGAAGCCGACGAAGAGACCGCCGUCCAGGAUCGUCUUUAAUGAGCCCUUGUAUUUUCACACAUCCCUAAACCACCCUCAUAUCGUGGCUGUGGUGGAAGUGGUCGCUGAGGGCAAGAAACGGGAUGGGAGCCUCCAGACAUUGUCCUGUGGGUUUGGAAUUCUUCGGAUCUUCAGCAACCAGCCGGACUCUCCUAUCUCUGCUUCCCAGGACAAAAGGUUGCGGCUGUACCAUGGCACCCCCAGAGCCCUCCUGCACCCGCUUCUCCAGGACCCCGCAGAGCAAAACAGACACAUGACCCUCAUUGAGAACUGCAGCCUGCAGUACACGCUGAAGCCACACCCGGCCCUGGAGCCUGCGUUCCACCUUCUUCCUGAGAACCUUCUGGUGUCUGGUCUGCAGCAGAUACCUGGCCUGCUUCCAGCUCAUGGAGAAUCCGAUCAUCCGGGAAGCCCACGGGACCCUCAGGCGGGCAGGAUGAACGACUGG | circ |
| ENSG00000131697:-:1:5961793:5986327 | ENSG00000131697 | ENST00000489180 | - | 1 | 5961794 | 5986327 | 22 | UGGAGAAUCCGAUCAUCCGGGA | bsj |
| ENSG00000131697:-:1:5961793:5986327 | ENSG00000131697 | ENST00000489180 | - | 1 | 5986318 | 5986527 | 210 | UUGAAAUGCAAAUGGUACUUCCUAGAGGAGGCAGCAUGGGUGCCAGCCAGCAACAAAGUCCACUCUUCUGGGGUUUGCAGAGUGUGGGUUUGGGGGAUGGAGGUUCCUCUGGGAUUUAGUGGGAGGCAGAAGCCUUCAGGAUUGCUGUGACCACUGUAGCCCUCACAGCUCUUCUCUUUAUUUGAAUACGACUGGCACAGAUCAUCCGGG | ie_up |
| ENSG00000131697:-:1:5961793:5986327 | ENSG00000131697 | ENST00000489180 | - | 1 | 5961594 | 5961803 | 210 | GGAGAAUCCGGUAAGGGCGUCUUUGAUUCUCUCCACUUGGUGAGGUGAGUCUGUCCUAGUUCACAGUGAAACCUUGAAGGCACUGGGCUCUUCUGGGGGCAGCGUGGGCCGAGCGCCCCUCUGCAGCUCAGCAGCACGGCCUGGCGGGGGUUGUGGGGGACAGUGGAGGAGGAUGGAAUGGCAGUCUAGUUUCCGUUUCCCUCCUAAAUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
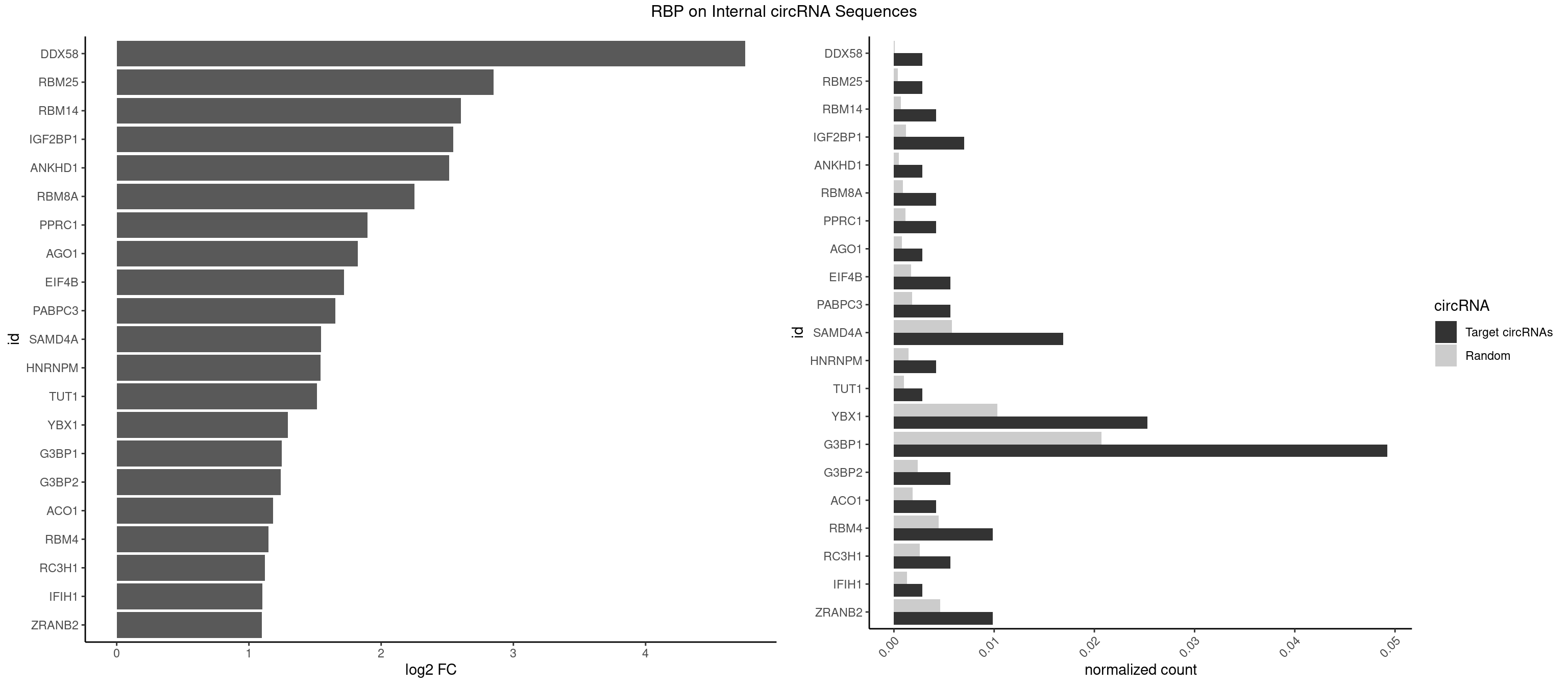
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| DDX58 | 1 | 71 | 0.002812940 | 0.0001043427 | 4.752677 | GCGCGC | GCGCGC |
| RBM25 | 1 | 268 | 0.002812940 | 0.0003898359 | 2.851140 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| RBM14 | 2 | 478 | 0.004219409 | 0.0006941687 | 2.603683 | CGCGCC,GCGCGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| IGF2BP1 | 4 | 831 | 0.007032349 | 0.0012057377 | 2.544091 | CACCCG,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ANKHD1 | 1 | 339 | 0.002812940 | 0.0004927293 | 2.513211 | GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| RBM8A | 2 | 611 | 0.004219409 | 0.0008869128 | 2.250177 | CGCGCC,GCGCGC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| PPRC1 | 2 | 780 | 0.004219409 | 0.0011318283 | 1.898386 | CGCGCC,GCGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| AGO1 | 1 | 548 | 0.002812940 | 0.0007956130 | 1.821940 | GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| EIF4B | 3 | 1179 | 0.005625879 | 0.0017100607 | 1.718031 | CUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPC3 | 3 | 1234 | 0.005625879 | 0.0017897669 | 1.652307 | AAAACA,AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SAMD4A | 11 | 3992 | 0.016877637 | 0.0057866714 | 1.544307 | CGGGAA,CGGGAC,CGGGCA,CUGGAC,CUGGCA,CUGGCC,CUGGUC,GCGGGC,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| HNRNPM | 2 | 999 | 0.004219409 | 0.0014492040 | 1.541780 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| TUT1 | 1 | 678 | 0.002812940 | 0.0009840095 | 1.515334 | AGAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| YBX1 | 17 | 7119 | 0.025316456 | 0.0103183321 | 1.294866 | AACCAC,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CAGCAA,CAUCUG,CCACAC,CCUGCG,GCCUGC,GGUCUG,GUCUGC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| G3BP1 | 34 | 14282 | 0.049226442 | 0.0206989801 | 1.249874 | ACACGC,ACCCAC,ACCCCC,ACCCGC,ACCGGC,AGGCAG,CACAGG,CACCCG,CAUCCG,CCACAC,CCACAG,CCACCC,CCCACA,CCCACC,CCCACG,CCCAGG,CCCCAC,CCCCAG,CCCCCA,CCCCGC,CCCGCA,CCCGGC,CCCUCC,CCGCAG,CCGCCG,CCGGCA,CCGGCC,CGGCAC,CGGCCC,UAGGCA | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| G3BP2 | 3 | 1644 | 0.005625879 | 0.0023839405 | 1.238730 | AGGAUG,GGAUGA,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ACO1 | 2 | 1283 | 0.004219409 | 0.0018607779 | 1.181135 | CAGUGA,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM4 | 6 | 3065 | 0.009845288 | 0.0044432593 | 1.147815 | CCUUCU,CUUCCU,CUUCUU,GCGCGC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| RC3H1 | 3 | 1786 | 0.005625879 | 0.0025897275 | 1.119278 | CCUUCU,CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| IFIH1 | 1 | 904 | 0.002812940 | 0.0013115296 | 1.100828 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| ZRANB2 | 6 | 3173 | 0.009845288 | 0.0045997733 | 1.097871 | AAAGGU,AGGUAC,CGGUAA,GAGGUU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
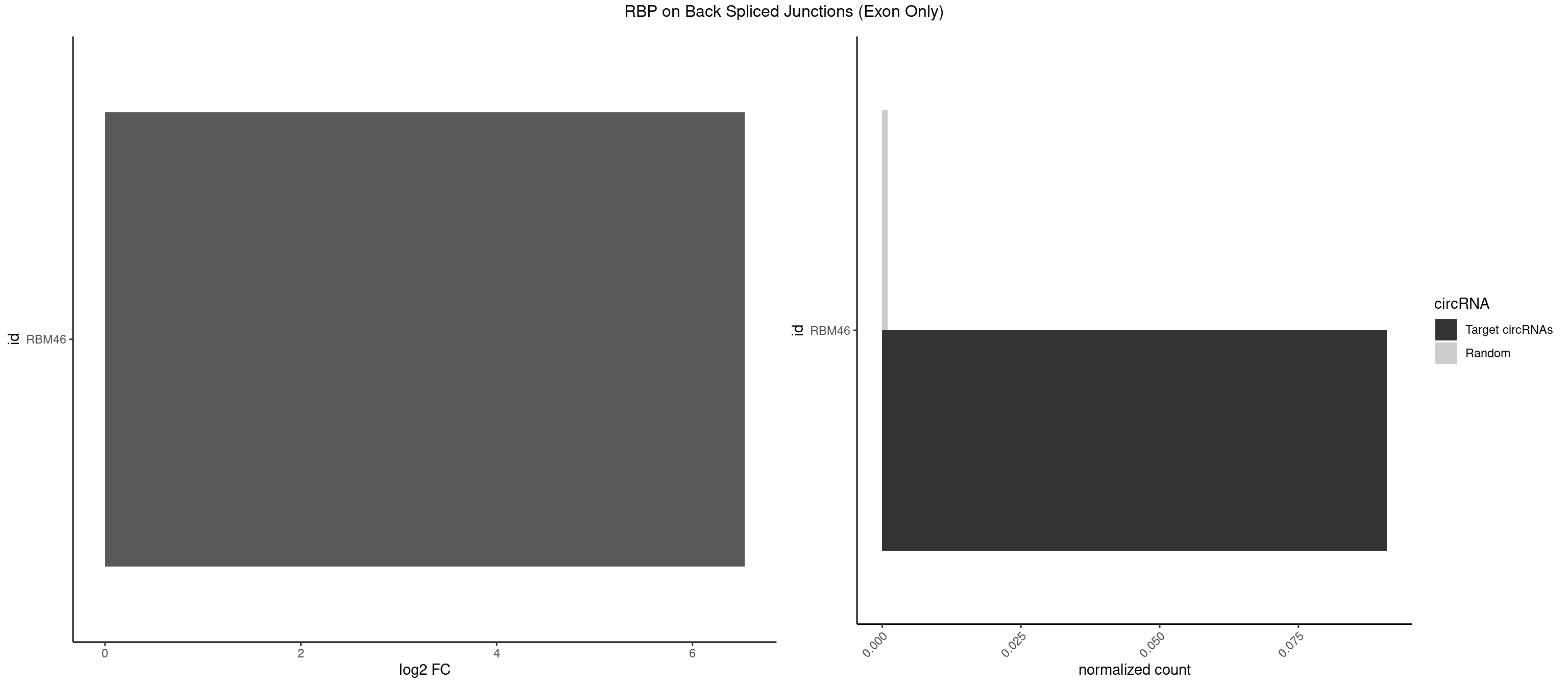
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM46 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | GAUCAU | AAUCAA,AAUGAU,AUCAAA,AUCAAU,AUGAAG,AUGAUG,AUGAUU,GAUCAU,GAUGAA,GAUGAU |
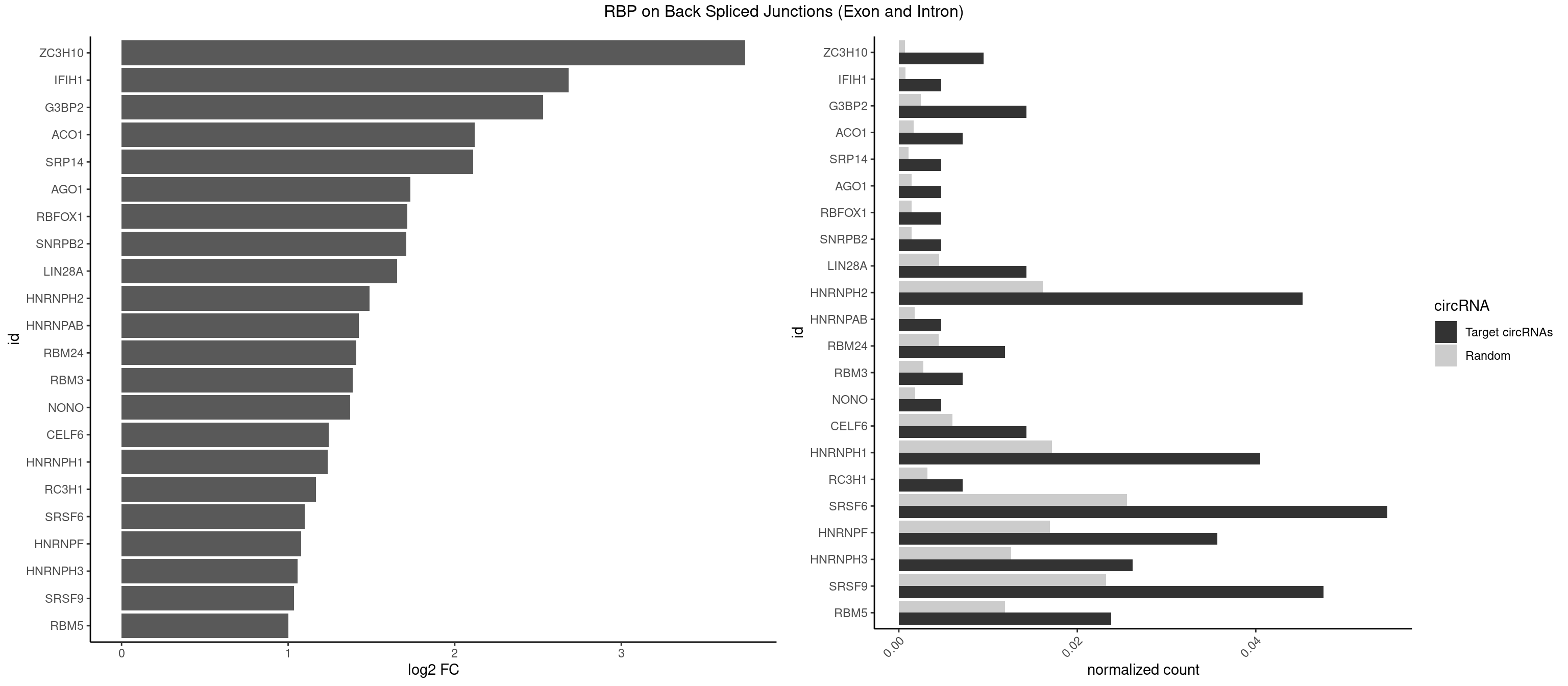
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZC3H10 | 3 | 140 | 0.009523810 | 0.0007103990 | 3.744837 | CGAGCG,GAGCGC,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| G3BP2 | 5 | 490 | 0.014285714 | 0.0024738009 | 2.529772 | AGGAUG,AGGAUU,GGAUGG,GGAUUG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| AGO1 | 1 | 283 | 0.004761905 | 0.0014308746 | 1.734641 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| LIN28A | 5 | 900 | 0.014285714 | 0.0045395002 | 1.653968 | GGAGAA,GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPH2 | 18 | 3198 | 0.045238095 | 0.0161174929 | 1.488911 | CGGGGG,CUGGGG,GGAAUG,GGAGGA,GGGAGG,GGGCGU,GGGGGA,GGGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | ACAAAG | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | AGAGUG,AGUGUG,GAGUGU,GUGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAUACG,AUACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| CELF6 | 5 | 1196 | 0.014285714 | 0.0060308343 | 1.244144 | GUGAGG,GUGGGG,GUGUGG,UGUGGG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPH1 | 16 | 3406 | 0.040476190 | 0.0171654575 | 1.237565 | CGGGGG,CUGGGG,GGAAUG,GGAGGA,GGGAGG,GGGCGU,GGGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CUUCUG | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| SRSF6 | 22 | 5069 | 0.054761905 | 0.0255441354 | 1.100181 | CACUGG,CAGCAC,CCACUC,CCACUG,CCUCAC,CCUCUG,CCUGGC,CUCUGG,CUUCAG,CUUCUC,CUUCUG,GCAGCA,UCACAG,UCAUCC,UCUCUC,UUCAGG,UUCUGG | AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
| HNRNPF | 14 | 3360 | 0.035714286 | 0.0169336961 | 1.076604 | CGGGGG,CUGGGG,GGAAUG,GGAGGA,GGAUGG,GGGAGG,GGGAUG,GGGGGC,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH3 | 10 | 2496 | 0.026190476 | 0.0125806127 | 1.057840 | CGGGGG,GGAAUG,GGAGGA,GGGAGG,GGGCGU,GGGGGC,UGGGUG,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| SRSF9 | 19 | 4608 | 0.047619048 | 0.0232214833 | 1.036079 | GAAGCC,GAAGGC,GAUGGA,GGACAG,GGAGAA,GGAGGA,GGAGGC,GGAUGG,GGGAGG,GGGUGC,GGUGCC,UGAAGG,UGACCA,UGGAGG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| RBM5 | 9 | 2359 | 0.023809524 | 0.0118903668 | 1.001746 | CUCUUC,CUUCUC,UCUUCU,UUCUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.