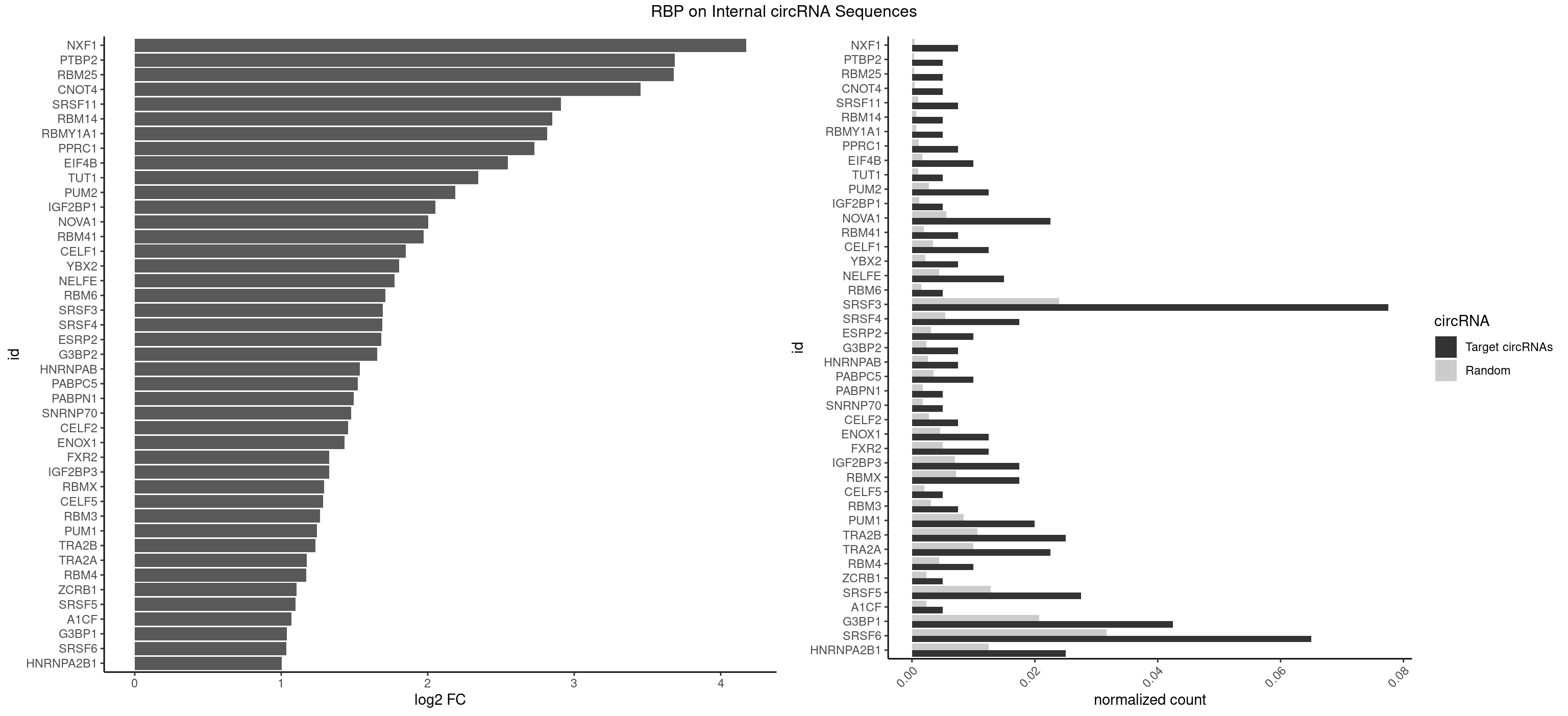
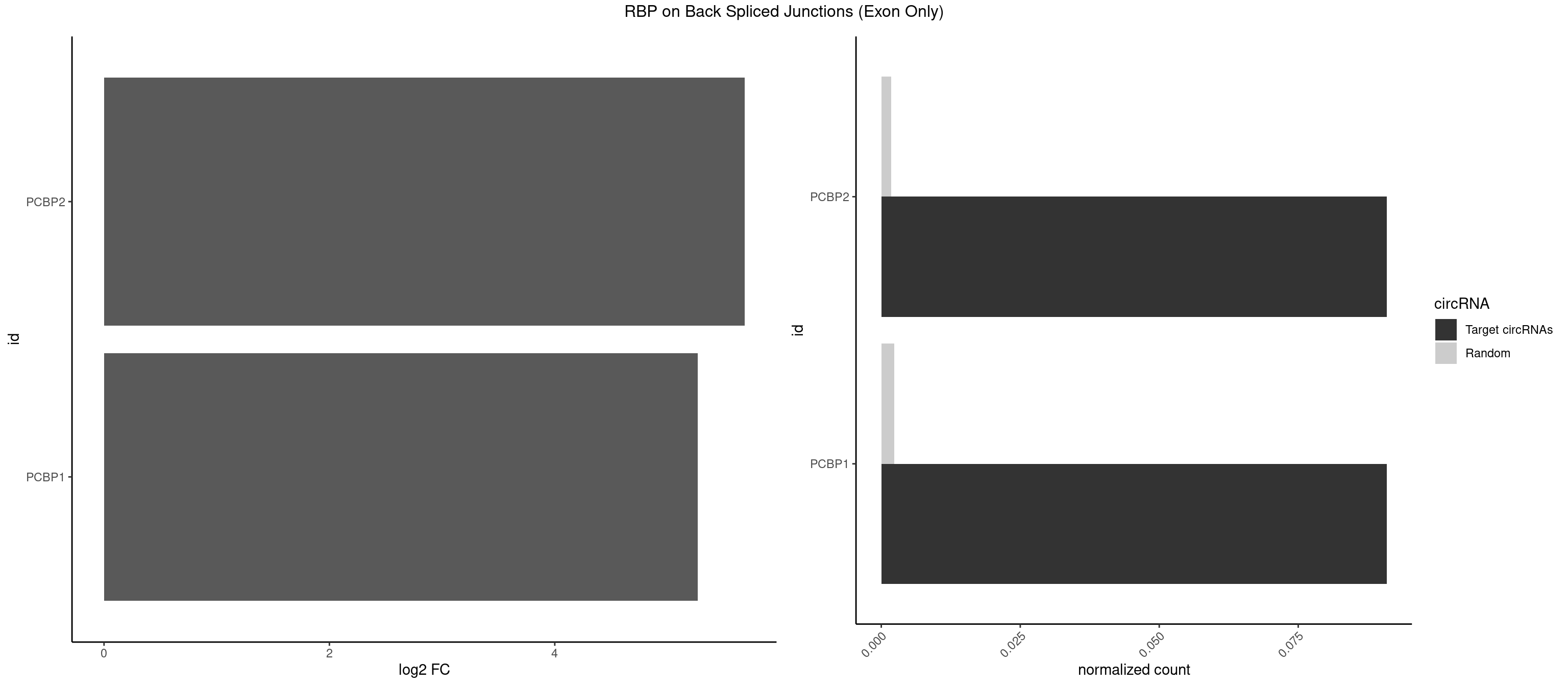
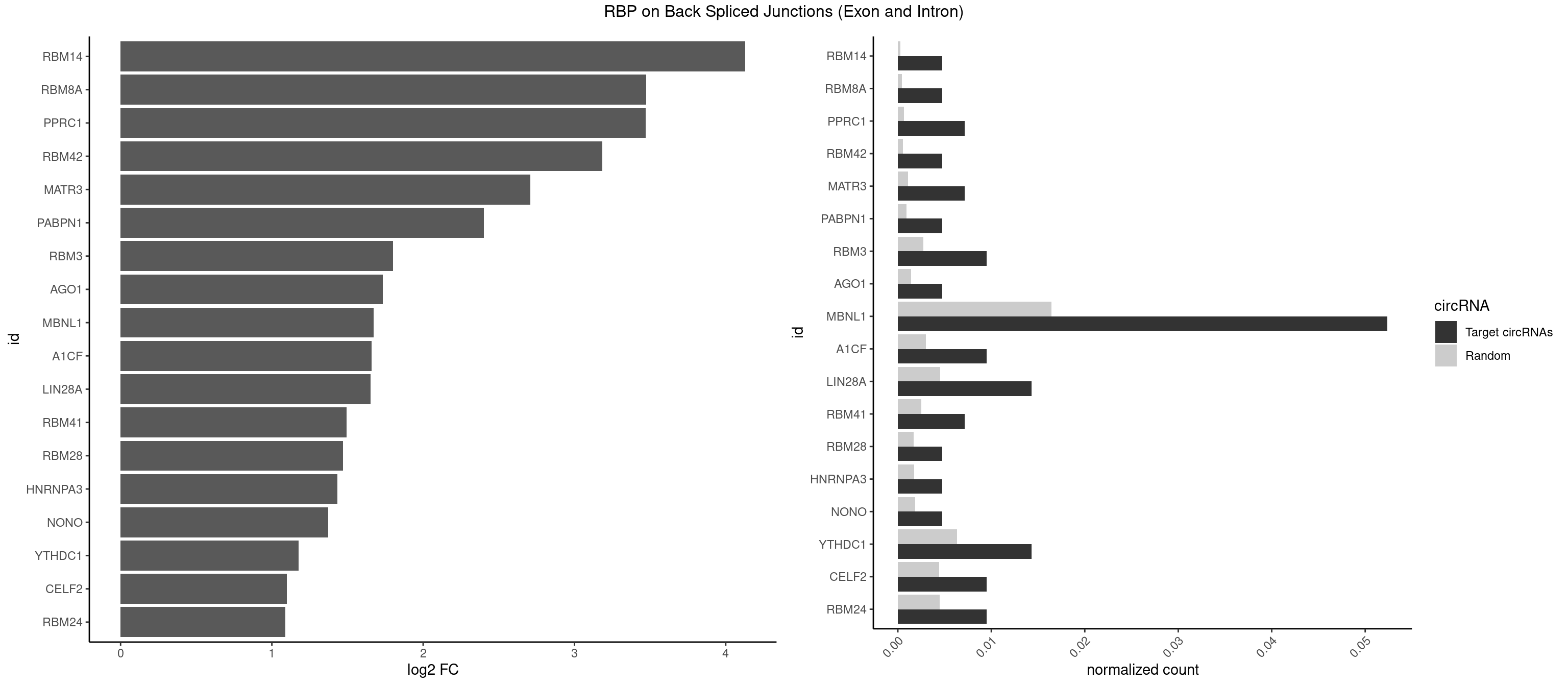
| NXF1 |
2 |
286 |
0.0075 |
0.0004159215 |
4.172507 |
AACCUG |
AACCUG |
| PTBP2 |
1 |
267 |
0.0050 |
0.0003883867 |
3.686363 |
CUCUCU |
CUCUCU |
| RBM25 |
1 |
268 |
0.0050 |
0.0003898359 |
3.680989 |
UCGGGC |
AUCGGG,CGGGCA,UCGGGC |
| CNOT4 |
1 |
314 |
0.0050 |
0.0004564992 |
3.453244 |
GACAGA |
GACAGA |
| SRSF11 |
2 |
688 |
0.0075 |
0.0009985015 |
2.909054 |
AAGAAG |
AAGAAG |
| RBM14 |
1 |
478 |
0.0050 |
0.0006941687 |
2.848570 |
GCGCGG |
CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBMY1A1 |
1 |
489 |
0.0050 |
0.0007101099 |
2.815814 |
ACAAGA |
ACAAGA,CAAGAC |
| PPRC1 |
2 |
780 |
0.0075 |
0.0011318283 |
2.728235 |
CGGCGC,GGCGCG |
CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| EIF4B |
3 |
1179 |
0.0100 |
0.0017100607 |
2.547881 |
CUUGGA,UUGGAA |
CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| TUT1 |
1 |
678 |
0.0050 |
0.0009840095 |
2.345184 |
AAAUAC |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| PUM2 |
4 |
1890 |
0.0125 |
0.0027404447 |
2.189446 |
GUACAU,UACAUC,UGUAAA,UGUACA |
GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| IGF2BP1 |
1 |
831 |
0.0050 |
0.0012057377 |
2.052012 |
GCACCC |
AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| NOVA1 |
8 |
3872 |
0.0225 |
0.0056127669 |
2.003141 |
AUCAAC,AUCAUC,AUUCAU,CAUUCA,UCAGUC,UCAUUC,UUCAUU |
AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| RBM41 |
2 |
1318 |
0.0075 |
0.0019115000 |
1.972185 |
AUACAU,UACAUU |
AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| CELF1 |
4 |
2391 |
0.0125 |
0.0034664959 |
1.850378 |
GUGUUG,GUUGUG,UGUUGU,UUGUGU |
CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| YBX2 |
2 |
1480 |
0.0075 |
0.0021462711 |
1.805058 |
ACAACU,ACAUCA |
AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| NELFE |
5 |
3028 |
0.0150 |
0.0043896388 |
1.772788 |
CUCUCU,CUCUGG,GUCUCU,UCUCUC,UCUCUG |
CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| RBM6 |
1 |
1054 |
0.0050 |
0.0015289102 |
1.709424 |
CAUCCA |
AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| SRSF3 |
30 |
16536 |
0.0775 |
0.0239654858 |
1.693238 |
ACAUCA,ACAUUC,ACUUCA,ACUUCG,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUUCAU,CACAUC,CAUCAA,CAUCAU,CAUCGA,CAUUCA,CCACCA,CUUCAA,CUUCAU,GACUUC,UACAUC,UACUUC,UCAUCC,UCAUCG,UCGACU,UUCAAC,UUCAUC |
AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| SRSF4 |
6 |
3740 |
0.0175 |
0.0054214720 |
1.690598 |
AAGAAG,AGAAGA,GAAGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| ESRP2 |
3 |
2150 |
0.0100 |
0.0031172377 |
1.681660 |
GGGAAG,GGGGAA,UGGGGA |
GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| G3BP2 |
2 |
1644 |
0.0075 |
0.0023839405 |
1.653542 |
AGGAUU,GGAUUA |
AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| HNRNPAB |
2 |
1782 |
0.0075 |
0.0025839306 |
1.537323 |
AGACAA,CAAAGA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PABPC5 |
3 |
2400 |
0.0100 |
0.0034795387 |
1.523032 |
AGAAAA,AGAAAU,GAAAAU |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| PABPN1 |
1 |
1222 |
0.0050 |
0.0017723764 |
1.496243 |
AGAAGA |
AAAAGA,AGAAGA |
| SNRNP70 |
1 |
1237 |
0.0050 |
0.0017941145 |
1.478656 |
AUUCAA |
AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF2 |
2 |
1886 |
0.0075 |
0.0027346479 |
1.455536 |
UGUUGU,UUGUGU |
AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ENOX1 |
4 |
3195 |
0.0125 |
0.0046316558 |
1.432328 |
AAUACA,AGACAG,CAGACA,UGUACA |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| FXR2 |
4 |
3434 |
0.0125 |
0.0049780156 |
1.328285 |
AGACAA,AGACAG,GACAGA,GGACGG |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| IGF2BP3 |
6 |
4815 |
0.0175 |
0.0069793662 |
1.326187 |
AAAUAC,AAUACA,ACAAUC,ACAUUC,CAUUCA |
AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| RBMX |
6 |
4925 |
0.0175 |
0.0071387787 |
1.293606 |
AAGAAG,AAGUGU,AGAAGG,AGUGUU,AUCCCC |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| CELF5 |
1 |
1415 |
0.0050 |
0.0020520728 |
1.284846 |
GUGUUG |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM3 |
2 |
2152 |
0.0075 |
0.0031201361 |
1.265282 |
AAUACG,AUACGA |
AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PUM1 |
7 |
5823 |
0.0200 |
0.0084401638 |
1.244657 |
GAAUUG,GUACAU,UAAUGU,UACAUC,UGUAAA,UGUACA,UGUCCA |
AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| TRA2B |
9 |
7329 |
0.0250 |
0.0106226650 |
1.234782 |
AAAGAA,AAGAAC,AAGAAG,AGAAGA,AGAAGG,GAAGAA |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| TRA2A |
8 |
6871 |
0.0225 |
0.0099589296 |
1.175862 |
AAAGAA,AAGAAA,AAGAAG,AGAAGA,GAAGAA,GAAGAG |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBM4 |
3 |
3065 |
0.0100 |
0.0044432593 |
1.170310 |
CGCGGU,CUUCCU,GCGCGG |
CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| ZCRB1 |
1 |
1605 |
0.0050 |
0.0023274215 |
1.103196 |
GGAUUA |
AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| SRSF5 |
10 |
8869 |
0.0275 |
0.0128544391 |
1.097165 |
AAGAAG,AGAAGA,CACAUC,CGCAUC,GAAGAA,GGAAGA,UAAAGG,UACAUC |
AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| A1CF |
1 |
1642 |
0.0050 |
0.0023810421 |
1.070335 |
AUCAGU |
AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| G3BP1 |
16 |
14282 |
0.0425 |
0.0206989801 |
1.037903 |
ACACGC,ACCCAC,ACCCCC,ACGCAC,CACCGG,CACGCA,CCACCC,CCACCG,CCAUCG,CCCACC,CCCAUC,CCCCCA,CCCCUA,UACGCC |
ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| SRSF6 |
25 |
21880 |
0.0650 |
0.0317100317 |
1.035500 |
AACCUG,AAGAAG,ACCGGG,ACCUGG,AGAAGA,AUCAAC,CAACCU,CAGCAC,CCUCAC,CCUGGC,CUCACG,CUCUGG,GAAGAA,GCACCU,GGAAGA,UCAACC,UCAUCC,UCUCUC,UCUCUG |
AACCUG,AAGAAG,ACCGGG,ACCGUC,ACCUGG,AGAAGA,AGCACC,AGCGGA,AGGAAG,AUCAAC,AUCCUG,AUCGUA,CAACCU,CACACG,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CAUCCU,CCACAC,CCACAG,CCACUC,CCACUG,CCCGGC,CCUCAC,CCUCAG,CCUCUC,CCUCUG,CCUGGC,CGCGUC,CUACAC,CUACAG,CUACUC,CUACUG,CUCACG,CUCAGG,CUCAUC,CUCUCG,CUCUGG,CUUCAC,CUUCAG,CUUCUC,CUUCUG,GAAGAA,GACGUC,GAGGAA,GAUCAA,GCACCU,GCAGCA,GCCGGA,GCCGUC,GCUCAU,GGAAGA,UACACG,UACAGG,UACGUC,UACUCG,UACUGG,UCAACC,UCACAC,UCACAG,UCACUC,UCACUG,UCAUCC,UCCGGA,UCCUGG,UCUCAC,UCUCAG,UCUCUC,UCUCUG,UGCGGA,UGCGGC,UGCGUA,UGCGUC,UGCGUG,UGUGGA,UUACAC,UUACAG,UUACUC,UUACUG,UUCACG,UUCAGG,UUCUCG,UUCUGG,UUUCAC,UUUCAG,UUUCUC,UUUCUG |
| HNRNPA2B1 |
9 |
8607 |
0.0250 |
0.0124747476 |
1.002917 |
AAGAAG,AGGGGC,CAAGAA,CCAAGA,GAAGCC,GCCAAG |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |