circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000075151:-:1:20941490:21111383
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000075151:-:1:20941490:21111383 | ENSG00000075151 | MSTRG.488.20 | - | 1 | 20941491 | 21111383 | 1941 | CAAUCCCACAGAGUAUUGAUGAGGAAACUGAAGUUUGGAGCGAUCACAUCAUUUUCCCAAGAAUUCCUAGAGGACCUGUGCAACAACCUCUUGAGGAUCGAAUCUUCACUCCCGCUGUCUCAGCAGUCUACAGCACGGUAACACAAGUGGCAAGACAGCCGGGAACCCCUACCCCAUCCCCUUAUUCAGCACAUGAAAUAAACAAGGGGCAUCCAAAUCUUGCGGCAACGCCCCCGGGACAUGCAUCGUCCCCUGGACUCUCUCAAACCCCUUAUCCCUCUGGACAGAAUGCAGGUCCAACCACGCUGGUAUACCCUCAAACCCCUCAGACAAUGAAUUCACAACCUCAAACCCGUUCUCCGCCCAGCAGAACAGUGCCAAUACAUUGCACAGACAACUGGAAGAGGAGGAAGGUUUUGGAACAGACUCCCGUUUACAGGUCCUUGGCUGGAAGAGGCUGGAUAAAAUACUGCAUUUUUUUCCAGAGGCCUCAAAUACAGCCUCCUAGAGCUACCAUCCCGAACAGCAGUCCUUCCAUUCGUCCUGGUGCACAGACACCCACUGCAGUGUACCAGGCUAAUCAGCACAUCAUGAUGGUUAACCAUCUGCCCAUGCCGUACCCAGUGCCCCAGGGGCCUCAGUACUGUAUACCACAGUACCGUCAUAGUGGCCCUCCUUAUGUUGGGCCCCCCCAACAAUAUCCAGUUCAACCACCGGGGCCAGGUCCUUUUUAUCCUGGACCAGGACCUGGGGACUUCCCCAAUGCUUAUGGAACGCCUUUUUACCCAAGUCAGCCGGUGUAUCAGUCAGCACCUAUCAUAGUGCCUACGCAGCAACAGCCGCCUCCAGCCAAGAGAGAGAAAAAAACUAUAAGAAUUCGGGAUCCAAACCAGGGAGGUAAAGACAUAACAGAGGAGAUUAUGUCUGGAGGUGGCAGCAGAAAUCCUACUCCACCCAUAGGAAGACCCACGUCCACACCUACUCCUCCUCAGCAGCUGCCCAGCCAGGUCCCCGAGCACAGCCCUGUGGUUUAUGGGACUGUGGAGAGCGCUCAUCUUGCUGCCAGCACCCCUGUCACUGCAGCUAGCGACCAGAAGCAAGAGGAGAAGCCAAAACCAGAUCCAGUGUUAAAGUCUCCUUCCCCAGUCCUUAGGCUAGUCCUCAGUGGAGAGAAGAAAGAACAAGAAGGCCAGACAUCUGAAACUACUGCAAUAGUAUCCAUAGCAGAGCUUCCUCUGCCUCCAUCACCUACCACUGUUUCUUCUGUUGCUCGAAGUACAAUUGCAGCCCCCACCUCUUCUGCUCUUAGUAGCCAACCAAUAUUCACCACUGCUAUAGAUGACAGAUGUGAACUCUCAUCCCCAAGAGAAGACACAAUUCCUAUACCCAGCCUCACAUCUUGCACAGAAACAUCAGACCCUUUACCAACAAAUGAAAAUGAUGAUGAUAUAUGCAAGAAACCCUGUAGUGUAGCACCUAAUGAUAUUCCACUGGUUUCUAGUACUAACCUAAUUAAUGAAAUAAAUGGAGUUAGCGAAAAAUUAUCAGCCACGGAGAGCAUUGUGGAAAUAGUAAAACAGGAAGUAUUGCCAUUGACUCUUGAAUUGGAGAUUCUCGAAAAUCCCCCAGAAGAAAUGAAACUGGAGUGUAUCCCAGCUCCCAUCACCCCUUCCACAGUUCCUUCCUUUCCUCCAACUCCUCCAACUCCUCCAGCUUCUCCUCCUCACACUCCAGUCAUUGUUCCUGCUGCUGCCACUACUGUUAGUUCUCCGAGUGCUGCCAUCACAGUCCAGAGAGUCCUAGAGGAGGACGAGAGCAUAAGAACUUGCCUUAGUGAAGAUGCAAAAGAGAUUCAGAACAAAAUAGAGGUAGAAGCAGAUGGGCAAACAGAAGAGAUUUUGGAUUCUCAAAACUUAAAUUCAAGAAGGAGCCCUGUCCCAGCAAUCCCACAGAGUAUUGAUGAGGAAACUGAAGUUUGGAGCGAUCACAUC | circ |
| ENSG00000075151:-:1:20941490:21111383 | ENSG00000075151 | MSTRG.488.20 | - | 1 | 20941491 | 21111383 | 22 | CCCUGUCCCAGCAAUCCCACAG | bsj |
| ENSG00000075151:-:1:20941490:21111383 | ENSG00000075151 | MSTRG.488.20 | - | 1 | 21111374 | 21111583 | 210 | AAGCUCUAAAGAAUGAAGUUAGGAGAUCCGUUGGUGCUCUUAAUUAGGAAUCUGGGAGAGAUGUCUUAAAGGCUGUGUCACCUGGAUAGUCCACGGAAAGGAAAACUCAUGAUCUUUAUGGAAGAGCUAUGUAUAAUAAUGAAAUACUGCUUAGUAUGUGCUAAGCCCAUCAUAAGCCUUAUUUUGCUAAUUCUUUCCAGCAAUCCCACA | ie_up |
| ENSG00000075151:-:1:20941490:21111383 | ENSG00000075151 | MSTRG.488.20 | - | 1 | 20941291 | 20941500 | 210 | CCUGUCCCAGGUAAGCCAUUCUGCCAUUAAUCUCGUGCUUGCUGAACAUGAAGAAGAGGCAGUGUUUGAGUUAUUUUUUAAUUGGCUACUUUCCUUGACUUCCAGAAACAUCGAAUGAAUGCUGAAAUUCCUAGUCUGGUUUCUUCAGUGGUGUGUUUGUGUUUUAGUACAACCUGCAAAAGAGUUACUCCAUUGUCUAAAAAUGCAAAU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
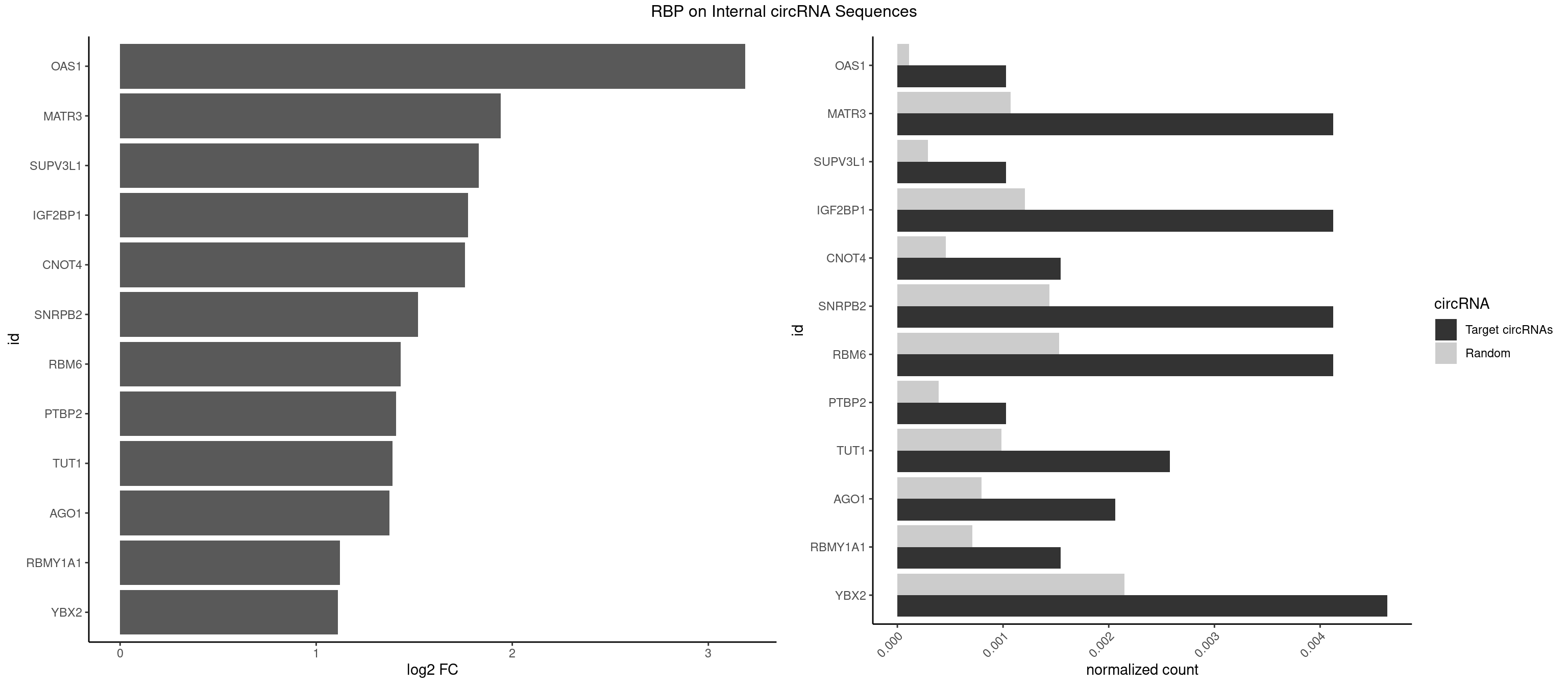
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.001030397 | 0.0001130379 | 3.188321 | GCAUAA | GCAUAA |
| MATR3 | 7 | 739 | 0.004121587 | 0.0010724109 | 1.942342 | AAUCUU,AUCUUG,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SUPV3L1 | 1 | 199 | 0.001030397 | 0.0002898408 | 1.829867 | CCGCCC | CCGCCC |
| IGF2BP1 | 7 | 831 | 0.004121587 | 0.0012057377 | 1.773284 | ACCCGU,AGCACC,CCCGUU,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| CNOT4 | 2 | 314 | 0.001545595 | 0.0004564992 | 1.759478 | GACAGA | GACAGA |
| SNRPB2 | 7 | 991 | 0.004121587 | 0.0014376103 | 1.519527 | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM6 | 7 | 1054 | 0.004121587 | 0.0015289102 | 1.430696 | AUCCAA,AUCCAG,CAUCCA,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| PTBP2 | 1 | 267 | 0.001030397 | 0.0003883867 | 1.407634 | CUCUCU | CUCUCU |
| TUT1 | 4 | 678 | 0.002575992 | 0.0009840095 | 1.388384 | AAAUAC,AAUACU,CAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| AGO1 | 3 | 548 | 0.002060793 | 0.0007956130 | 1.373061 | AGGUAG,GAGGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| RBMY1A1 | 2 | 489 | 0.001545595 | 0.0007101099 | 1.122048 | ACAAGA,CAAGAC | ACAAGA,CAAGAC |
| YBX2 | 8 | 1480 | 0.004636785 | 0.0021462711 | 1.111293 | AACAAC,AACAUC,ACAACU,ACAUCA,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
RBP on BSJ (Exon Only)
Plot
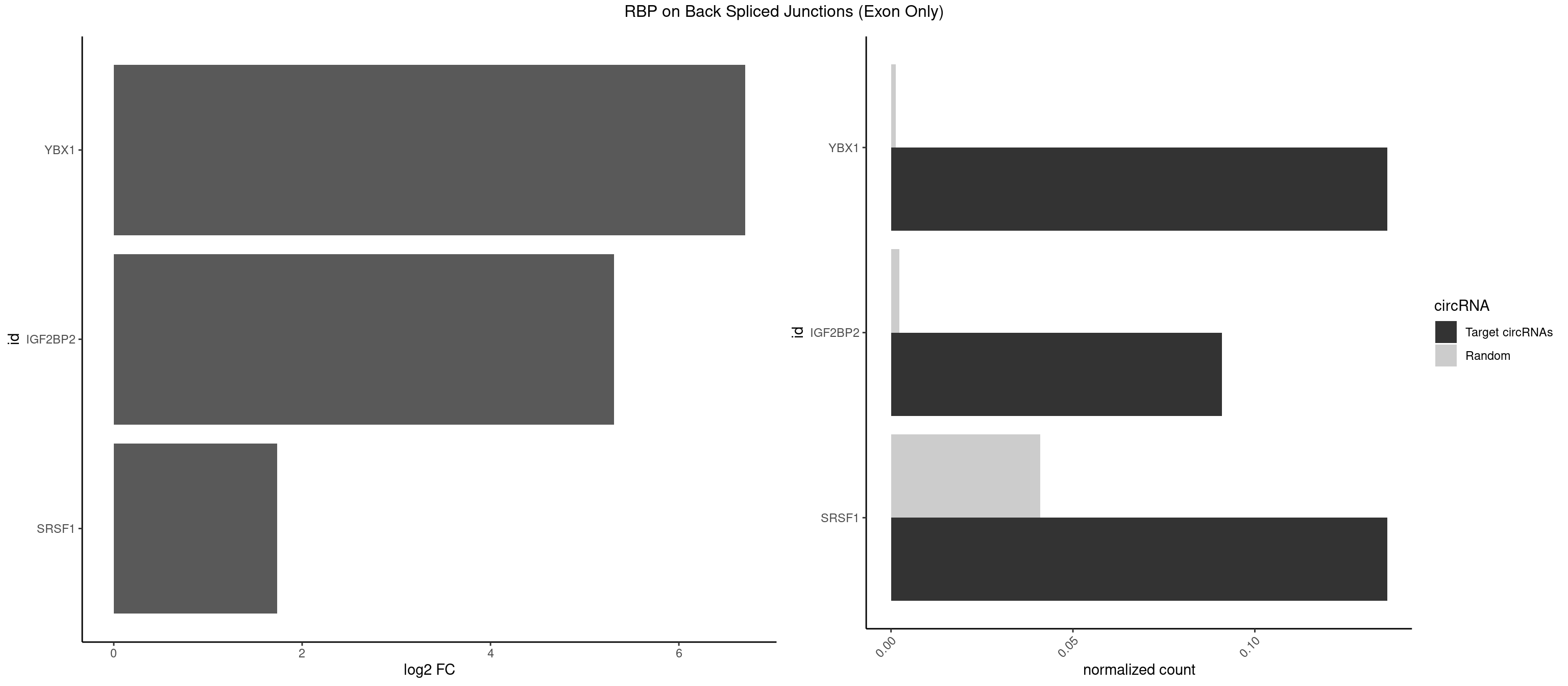
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| YBX1 | 2 | 19 | 0.13636364 | 0.001309929 | 6.701826 | CAGCAA,CCAGCA | AACCAC,ACACCA,ACAUCU,ACCACC,CACCAC,CAGCAA,CCACCA,CCAGCA,CCUGCG,CUGCGG,GAUCUG,GCCUGC,UCCAGC,UGCGGU |
| IGF2BP2 | 1 | 34 | 0.09090909 | 0.002292376 | 5.309509 | GCAAUC | AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,ACAAAC,ACAAUC,ACACUC,ACAUAC,CAAAAC,CAAUAC,CAAUCA,CAAUUC,CACUCA,CAUACA,CAUUCA,CCAUAC,CCAUUC,GAAAAC,GAACAC,GAAUAC,GAAUUC |
| SRSF1 | 2 | 625 | 0.13636364 | 0.041000786 | 1.733736 | CCAGCA,CCCAGC | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
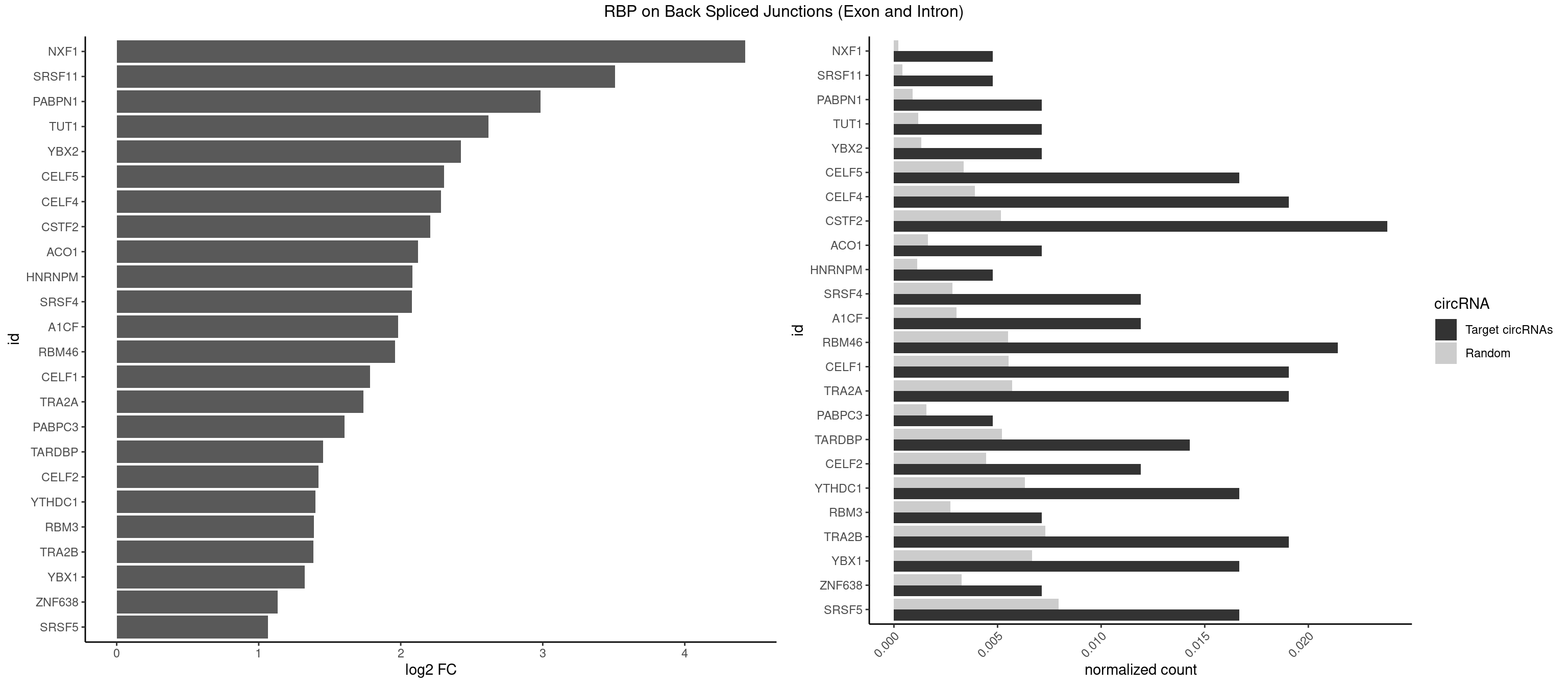
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| TUT1 | 2 | 230 | 0.007142857 | 0.0011638452 | 2.617602 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| YBX2 | 2 | 263 | 0.007142857 | 0.0013301088 | 2.424957 | AACAUC,ACAUCG | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CELF5 | 6 | 669 | 0.016666667 | 0.0033756550 | 2.303726 | GUGUGU,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF4 | 7 | 776 | 0.019047619 | 0.0039147521 | 2.282618 | GGUGUG,GUGUGU,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CSTF2 | 9 | 1022 | 0.023809524 | 0.0051541717 | 2.207726 | GUGUGU,GUGUUU,UGUGUU,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGG,CAGUGU | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| SRSF4 | 4 | 558 | 0.011904762 | 0.0028164047 | 2.079612 | AAGAAG,AGAAGA,GAAGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| A1CF | 4 | 598 | 0.011904762 | 0.0030179363 | 1.979904 | UAAUUA,UAAUUG,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| RBM46 | 8 | 1091 | 0.021428571 | 0.0055018138 | 1.961556 | AAUGAA,AUCAUA,AUGAAA,AUGAAG,AUGAAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| CELF1 | 7 | 1097 | 0.019047619 | 0.0055320435 | 1.783726 | GUGUGU,GUUUGU,UGUGUU,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| TRA2A | 7 | 1133 | 0.019047619 | 0.0057134220 | 1.737184 | AAAGAA,AAGAAG,AAGAGG,AGAAGA,GAAGAA,GAAGAG | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| TARDBP | 5 | 1034 | 0.014285714 | 0.0052146312 | 1.453936 | GAAUGA,GUGUGU,UGAAUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| CELF2 | 4 | 881 | 0.011904762 | 0.0044437727 | 1.421682 | GUAUGU,GUGUGU,UAUGUG,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| YTHDC1 | 6 | 1254 | 0.016666667 | 0.0063230552 | 1.398272 | GAAUGC,GGCUAC,UACUGC,UAGUAC,UCGUGC,UGGUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,AAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| TRA2B | 7 | 1447 | 0.019047619 | 0.0072954454 | 1.384543 | AAAGAA,AAGAAG,AAGAAU,AAGGAA,AGAAGA,AGGAAA,GAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| YBX1 | 6 | 1321 | 0.016666667 | 0.0066606207 | 1.323237 | AACAUC,ACAUCG,CAGCAA,CAUCAU,CCAGCA,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | CGUUGG,GUUGGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| SRSF5 | 6 | 1578 | 0.016666667 | 0.0079554615 | 1.066948 | AAGAAG,AGAAGA,CACGGA,GAAGAA,GGAAGA,UAAAGG | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.