circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000196365:-:19:5705771:5714271
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000196365:-:19:5705771:5714271 | ENSG00000196365 | ENST00000360614 | - | 19 | 5705772 | 5714271 | 938 | GUUAAAAAUAAGAAGUUGGUUGAGCUGCUGAGAAGGAAAGUUCGUCUCGCCCAGCCUUAUGUCGGCGUCUUUCUAAAGAGAGAUGACAGCAAUGAGUCGGAUGUGGUCGAGAGCCUGGAUGAAAUCUACCACACGGGGACGUUUGCCCAGAUCCAUGAGAUGCAGGACCUUGGGGACAAGCUGCGCAUGAUCGUCAUGGGACACAGAAGAGUCCAUAUCAGCAGACAGCUGGAGGUGGAGCCCGAGGAGCCGGAGGCGGAGAACAAGCACAAGCCCCGCAGGAAGUCAAAGCGGGGCAAGAAGGAGGCGGAGGACGAGCUGAGCGCCAGGCACCCGGCGGAGCUGGCGAUGGAGCCCACCCCUGAGCUCCCGGCUGAGGUGCUCAUGGUGGAGGUAGAGAACGUUGUCCACGAGGACUUCCAGGUCACGGAGGAGGUGAAAGCCCUGACUGCAGAGAUCGUGAAGACCAUCCGGGACAUCAUUGCCUUGAACCCUCUCUACAGGGAGUCAGUGCUGCAGAUGAUGCAGGCUGGCCAGCGGGUGGUGGACAACCCCAUCUACCUGAGCGACAUGGGCGCCGCGCUCACCGGGGCCGAGUCCCAUGAGCUGCAGGACGUCCUGGAAGAGACCAAUAUUCCUAAGCGGCUGUACAAGGCCCUCUCCCUGCUGAAGAAGGAAUUUGAACUGAGCAAGCUGCAGCAGCGCCUGGGGCGGGAGGUGGAGGAGAAGAUCAAGCAGACCCACCGUAAGUACCUGCUGCAGGAGCAGCUAAAGAUCAUCAAGAAGGAGCUGGGCCUGGAGAAGGACGACAAGGAUGCCAUCGAGGAGAAGUUCCGGGAGCGCCUGAAGGAGCUCGUGGUCCCCAAGCACGUCAUGGAUGUUGUGGACGAGGAGCUGAGCAAGCUGGGCCUGCUGGACAACCACUCCUCGGAGUUCAAGUUAAAAAUAAGAAGUUGGUUGAGCUGCUGAGAAGGAAAGUUCGUCUCGC | circ |
| ENSG00000196365:-:19:5705771:5714271 | ENSG00000196365 | ENST00000360614 | - | 19 | 5705772 | 5714271 | 22 | UCGGAGUUCAAGUUAAAAAUAA | bsj |
| ENSG00000196365:-:19:5705771:5714271 | ENSG00000196365 | ENST00000360614 | - | 19 | 5714262 | 5714471 | 210 | UUAGCCAGGUGUGGUGGUGCAUGCCUGUAAUCCCAGCUACUCAGGAGGCUGAGGCAGGAGAAUCCCUUGAACUCAGUGGGGGUGGAGGUUGCAGUGAGCUGAGAUCACGCCAUUACAUUCCAGCCAGGGCAACAGAAUGAGACUCUGUCUCAAAAAAAAAAUCUACUCUGCAUUUCACUUUGGGGUCAUUGUCAUGCUAGGUUAAAAAUA | ie_up |
| ENSG00000196365:-:19:5705771:5714271 | ENSG00000196365 | ENST00000360614 | - | 19 | 5705572 | 5705781 | 210 | CGGAGUUCAAGUGAGUGCCAGGCCCGGGGCGGCCCAGCCCUCAGGUGACCCCUCCUCAGCCCCCAUCUUAGGCAGGCCAGGGGACCCAGCGGGGAGCCCCGUGGCACCUUCUUGGUGUGGCAGGUGCGGGCUGGGAGGCAUCCAGCGAGUAUCAGCCCUGUUCUGGGCAGGACACAUCAUCACCAUCUCCAUUCUACAAGGAAAUCGAGG | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
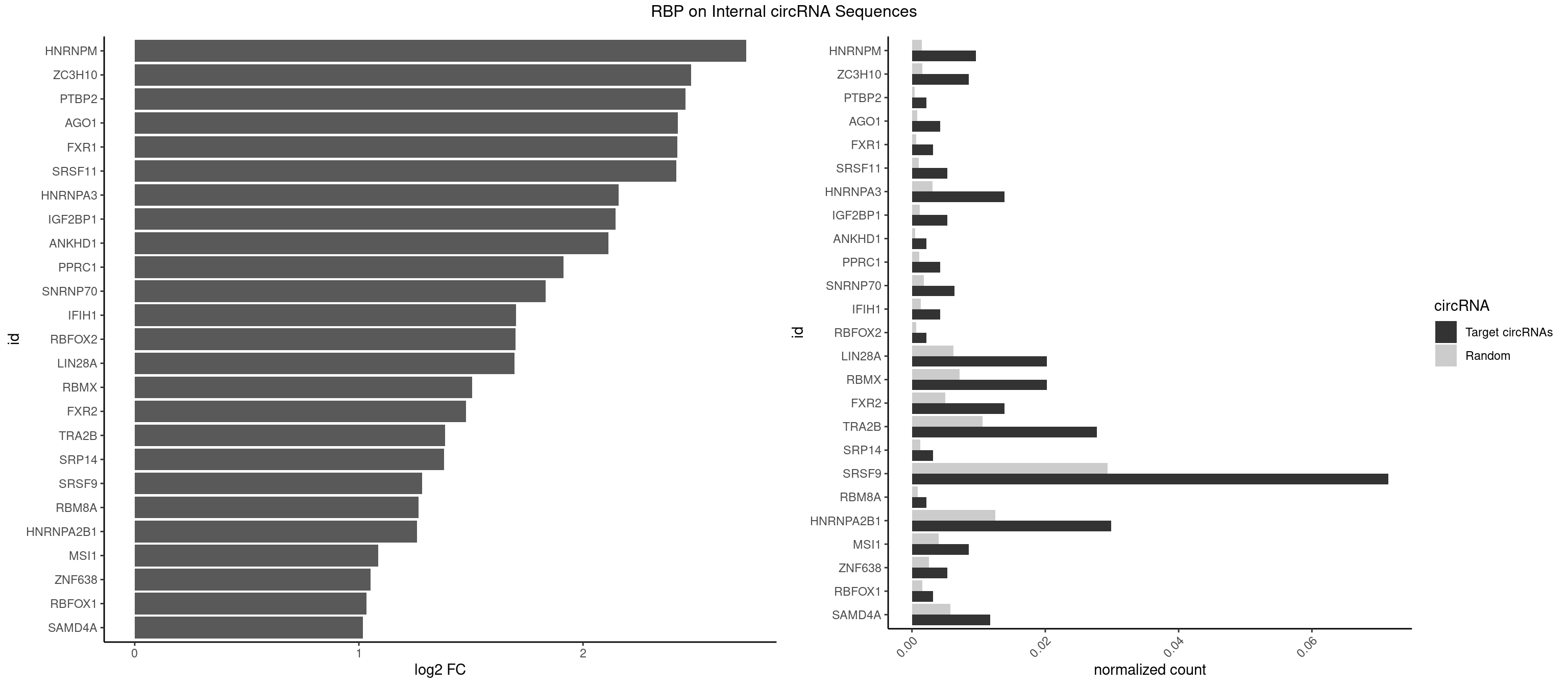
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPM | 8 | 999 | 0.009594883 | 0.0014492040 | 2.727005 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ZC3H10 | 7 | 1053 | 0.008528785 | 0.0015274610 | 2.481205 | CAGCGC,CCAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| PTBP2 | 1 | 267 | 0.002132196 | 0.0003883867 | 2.456775 | CUCUCU | CUCUCU |
| AGO1 | 3 | 548 | 0.004264392 | 0.0007956130 | 2.422201 | AGGUAG,GAGGUA,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| FXR1 | 2 | 411 | 0.003198294 | 0.0005970720 | 2.421326 | ACGACA,AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRSF11 | 4 | 688 | 0.005330490 | 0.0009985015 | 2.416432 | AAGAAG | AAGAAG |
| HNRNPA3 | 12 | 2140 | 0.013859275 | 0.0031027457 | 2.159234 | AAGGAG,AGGAGC,CAAGGA,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| IGF2BP1 | 4 | 831 | 0.005330490 | 0.0012057377 | 2.144352 | AAGCAC,CACCCG,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| ANKHD1 | 1 | 339 | 0.002132196 | 0.0004927293 | 2.113473 | GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| PPRC1 | 3 | 780 | 0.004264392 | 0.0011318283 | 1.913685 | CCGCGC,GGCGCC,GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| SNRNP70 | 5 | 1237 | 0.006396588 | 0.0017941145 | 1.834031 | AUCAAG,GAUCAA,GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| IFIH1 | 3 | 904 | 0.004264392 | 0.0013115296 | 1.701090 | GCCGCG,GGCCCU,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| RBFOX2 | 1 | 452 | 0.002132196 | 0.0006564894 | 1.699497 | UGACUG | UGACUG,UGCAUG |
| LIN28A | 18 | 4315 | 0.020255864 | 0.0062547643 | 1.695312 | AGGAGA,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGGA,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| RBMX | 18 | 4925 | 0.020255864 | 0.0071387787 | 1.504590 | AAGAAG,AAGGAA,AGAAGG,AGGAAG,GAAGGA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| FXR2 | 12 | 3434 | 0.013859275 | 0.0049780156 | 1.477209 | AGACAG,GACAAG,GACGAG,GGACAA,GGACGA,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| TRA2B | 25 | 7329 | 0.027718550 | 0.0106226650 | 1.383706 | AAGAAG,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAGAA,GAAGGA,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SRP14 | 2 | 847 | 0.003198294 | 0.0012289250 | 1.379906 | CGCCUG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| SRSF9 | 66 | 20254 | 0.071428571 | 0.0293536261 | 1.282963 | AGGAAA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGACA,AUGAGA,AUGAGC,CUGGAU,GAAGGA,GAUGCA,GAUGCC,GAUGGA,GGAAAG,GGACAA,GGAGAA,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGGAGC,GGGAGG,GGGUGG,GGUGGA,UGAAAG,UGAAGG,UGACAG,UGAGAA,UGAGCA,UGAGCG,UGAUGC,UGGAGC,UGGAGG,UGGUGG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| RBM8A | 1 | 611 | 0.002132196 | 0.0008869128 | 1.265476 | CGCGCU | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| HNRNPA2B1 | 27 | 8607 | 0.029850746 | 0.0124747476 | 1.258756 | AAGAAG,AAGGAA,AAGGAG,AGGAGC,CAAGAA,CAAGGA,GAAGGA,GGAGCC,GGGGCC | AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| MSI1 | 7 | 2770 | 0.008528785 | 0.0040157442 | 1.086673 | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUUGG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| ZNF638 | 4 | 1773 | 0.005330490 | 0.0025708878 | 1.052002 | CGUUGU,GUUCGU,GUUGGU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBFOX1 | 2 | 1077 | 0.003198294 | 0.0015622419 | 1.033685 | GCAUGA,UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SAMD4A | 10 | 3992 | 0.011727079 | 0.0057866714 | 1.019038 | CGGGAC,CUGGAA,CUGGAC,CUGGCC,GCGGGA,GCGGGU,GCUGGA,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
RBP on BSJ (Exon Only)
Plot
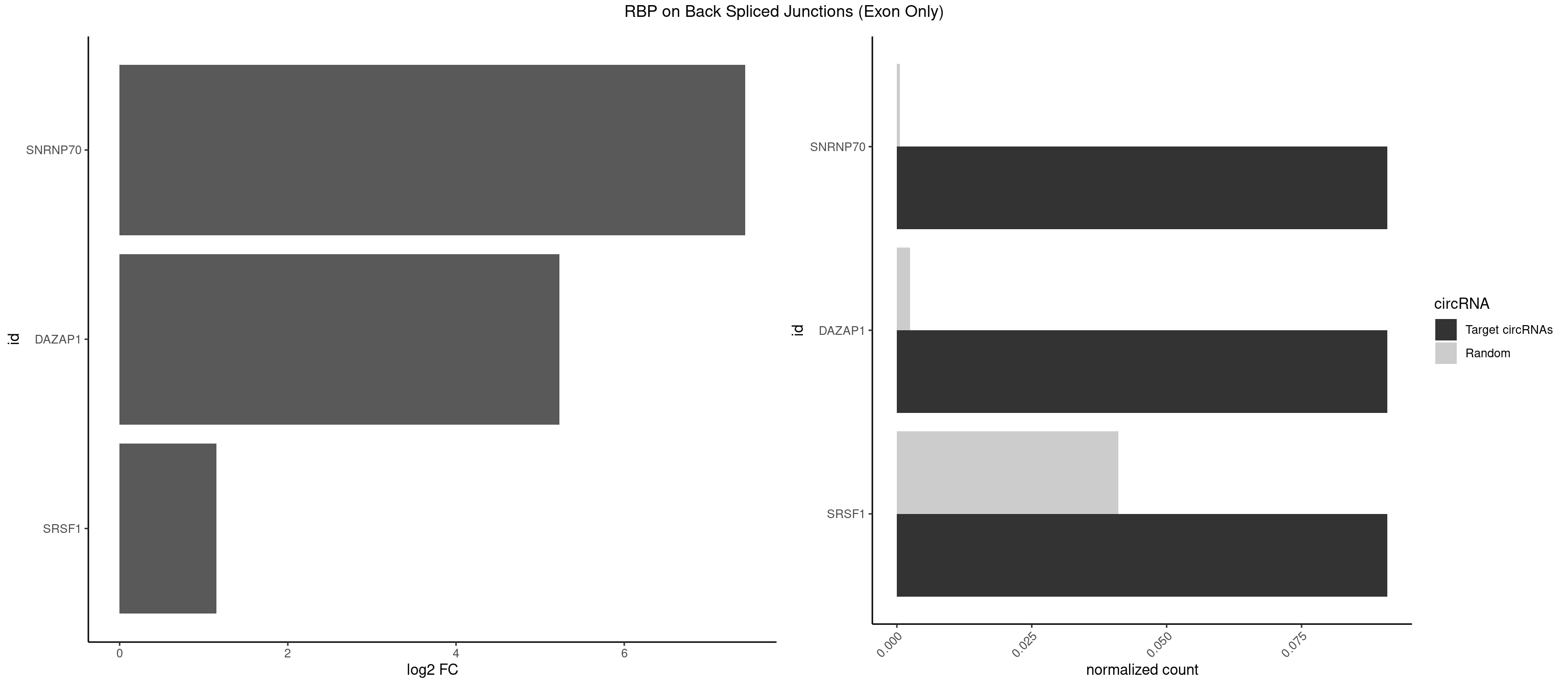
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRNP70 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | UUCAAG | AAUCAA,AUUCAA,GUUCAA,UUCAAG |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.0024233691 | 5.229338 | AGUUAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
| SRSF1 | 1 | 625 | 0.09090909 | 0.0410007860 | 1.148773 | UUCAAG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AACAGC,AAGAAC,AAGACC,AAGAGA,AAGAGC,AAGAGG,AAGGAC,AAGGCG,AAGGUG,AAUGAC,AAUUUC,ACAAGG,ACAGAG,ACAGCA,ACAGCG,ACAGGA,ACAGGG,ACAGGU,ACAGUG,ACCCGA,ACCGGA,ACGAAU,ACGGAA,ACUGAG,ACUGGA,AGAAGA,AGAAGG,AGACAA,AGACAG,AGACGU,AGAGAA,AGAGAC,AGAGCA,AGAGGA,AGAGGG,AGAGGU,AGAUGG,AGCAGG,AGCCGA,AGCGGA,AGGAAA,AGGAAC,AGGAAG,AGGACA,AGGACC,AGGACG,AGGACU,AGGAGA,AGGAGC,AGGAGG,AGGUAA,AUGAAC,AUGAAG,AUGACA,AUGACU,AUGGAC,AUGGAG,CAAGGA,CAAUGG,CACAGA,CACAGC,CACAGG,CACAGU,CACCCA,CACCCG,CACCGG,CACGCA,CACGGA,CACUGG,CAGAAC,CAGACA,CAGACG,CAGAGA,CAGAGC,CAGAGG,CAGCCG,CAGUCG,CAUGGU,CCAACC,CCAAGG,CCACCA,CCACCC,CCACGG,CCAGCA,CCAGCC,CCAGCG,CCAGGA,CCAGGG,CCCACC,CCCAGC,CCCAGG,CCCCGC,CCCGGG,CCCGUU,CCCUCC,CCCUCG,CCGAGG,CCGCGA,CCGCUA,CCGGAC,CCGGAG,CCGGGA,CCGUCC,CCGUGC,CCGUGG,CCGUUU,CCUAGG,CCUCCG,CCUCGA,CCUGCG,CCUGGA,CCUGGG,CGAACG,CGAAGC,CGAGGA,CGAGGC,CGAGGG,CGAUGG,CGCAGC,CGCCGC,CGCUAU,CGCUGC,CGGAAU,CGGACA,CGGAGC,CGGAGG,CGGCGG,CGGGCA,CGGUGC,CGGUGG,CGUGCG,CGUGGA,CGUGGG,CUCAGG,CUCGUG,CUGAAC,CUGAGU,GAAAGA,GAAAGG,GAACAG,GAAGAA,GAAGAG,GAAGAU,GAAGCA,GAAGCC,GAAGCU,GAAGGA,GAAGGC,GAAGGU,GAAUGA,GACAGA,GACAGG,GACCCA,GACGAA,GACGAC,GACGGA,GACUGA,GAGAAC,GAGAAG,GAGACA,GAGACG,GAGCAG,GAGGAA,GAGGAC,GAGGAG,GAGGAU,GAGGCA,GAGGGA,GAGGGC,GAGGGG,GAGGUA,GAUGAA,GAUGAC,GAUGAU,GAUGCA,GAUGCC,GAUGCU,GAUGGA,GAUGGC,GCACGG,GCAGCA,GCAGCG,GCAGGA,GCAGGC,GCAGGG,GCAGGU,GCCCAC,GCCCGG,GCCCGU,GCGCAA,GCGCCA,GCGCCC,GCGCGG,GCGGAC,GCGGCG,GCGGUU,GCUGGG,GGAAAG,GGAACA,GGAAGA,GGAAGG,GGAAUG,GGACAA,GGACAG,GGACCA,GGACCG,GGACGA,GGAGAA,GGAGAC,GGAGAU,GGAGCA,GGAGCG,GGAGCU,GGAGGA,GGAGGC,GGAGGG,GGAGGU,GGAUAU,GGAUGA,GGAUUC,GGCACA,GGCAGA,GGCCGA,GGCGCA,GGGACG,GGGCCG,GGGGAA,GGGGAG,GGGGCA,GGGGCG,GGGGGA,GGGUAC,GGUCCA,GGUCCG,GGUGAA,GGUGCA,GGUGCC,GGUGCG,GGUGCU,GGUGGA,GGUGGC,GGUGGG,GGUGGU,GUAGGA,GUGACA,UAGACA,UAGGAC,UCAAGA,UCAGGU,UCGGGC,UGAACA,UGAAGA,UGAAGC,UGAAGG,UGACAG,UGACGA,UGACUG,UGAGUU,UGAUGA,UGAUGG,UGGACA,UGGAGA,UGGAGC,UGGAGG,UGGUGC,UGUAGG,UUCAAG |
RBP on BSJ (Exon and Intron)
Plot
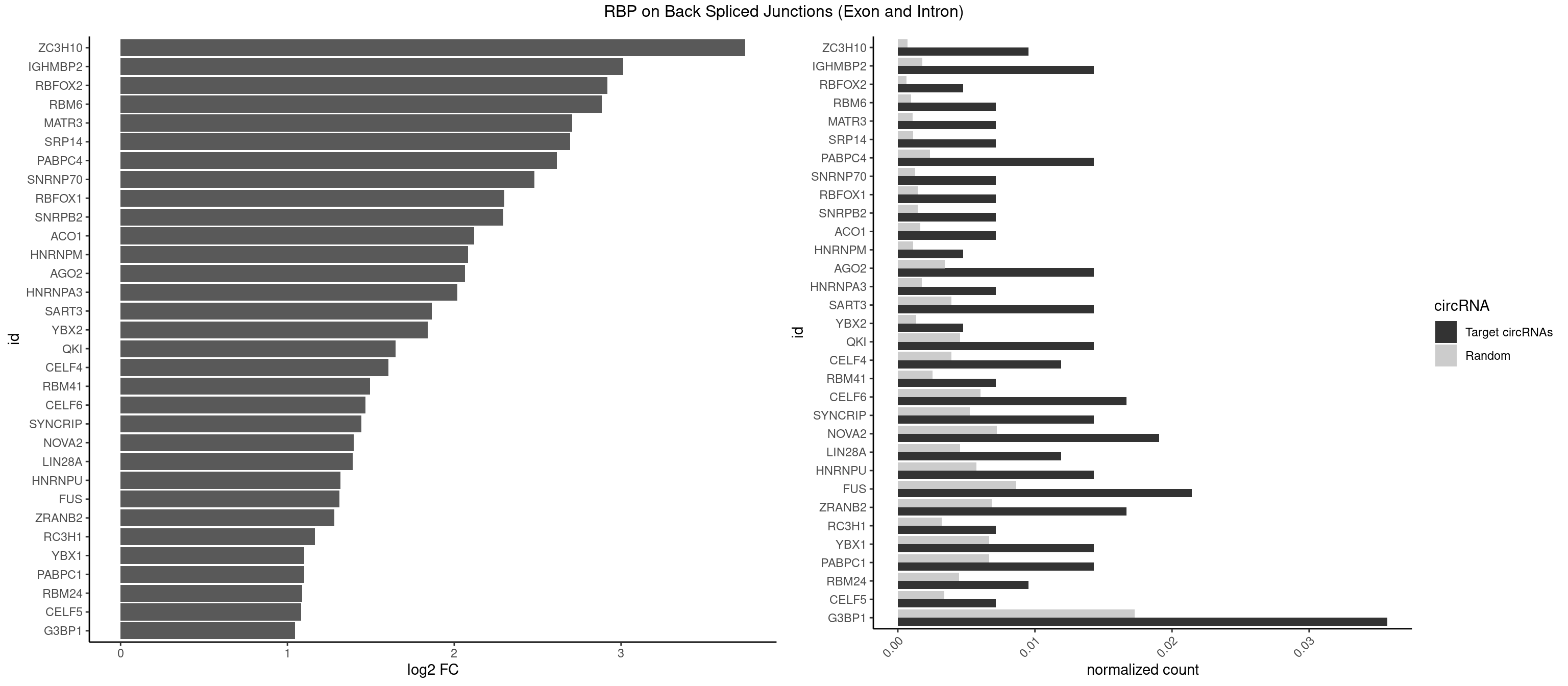
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ZC3H10 | 3 | 140 | 0.009523810 | 0.0007103990 | 3.744837 | CAGCGA,CCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| IGHMBP2 | 5 | 350 | 0.014285714 | 0.0017684401 | 3.014024 | AAAAAA | AAAAAA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AUCCAG,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| PABPC4 | 5 | 462 | 0.014285714 | 0.0023327287 | 2.614483 | AAAAAA | AAAAAA,AAAAAG |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.0012797259 | 2.480666 | GUUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.0014510278 | 2.299426 | GCAUGC,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | AAGGAA | AAGGAA,GAAGGA,GGGGGG |
| AGO2 | 5 | 677 | 0.014285714 | 0.0034159613 | 2.064210 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | CAAGGA,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| SART3 | 5 | 777 | 0.014285714 | 0.0039197904 | 1.865725 | AAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| YBX2 | 1 | 263 | 0.004761905 | 0.0013301088 | 1.839994 | ACAUCA | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| QKI | 5 | 904 | 0.014285714 | 0.0045596534 | 1.647577 | AUCUAC,CUACUC,UACUCA,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| CELF4 | 4 | 776 | 0.011904762 | 0.0039147521 | 1.604546 | GGUGUG,GUGUGG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| RBM41 | 2 | 502 | 0.007142857 | 0.0025342604 | 1.494937 | UACAUU,UUACAU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| CELF6 | 6 | 1196 | 0.016666667 | 0.0060308343 | 1.466536 | GUGGGG,GUGGUG,GUGUGG,UGUGGU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| SYNCRIP | 5 | 1041 | 0.014285714 | 0.0052498992 | 1.444212 | AAAAAA | AAAAAA,UUUUUU |
| NOVA2 | 7 | 1434 | 0.019047619 | 0.0072299476 | 1.397554 | AGAUCA,AGGCAU,AUCACC,AUCAUC,GAGGCA,GAUCAC | AACACC,ACCACC,AGACAU,AGAUCA,AGCACC,AGGCAU,AGUCAU,AUCAAC,AUCACC,AUCAUC,CCUAGA,CUAGAU,GAGACA,GAGGCA,GAGUCA,GAUCAC,GGGGGG,UAGAUC,UUUUUU |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | AGGAGA,CGGAGU,GGAGAA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| HNRNPU | 5 | 1136 | 0.014285714 | 0.0057285369 | 1.318335 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| FUS | 8 | 1711 | 0.021428571 | 0.0086255542 | 1.312847 | AAAAAA,GGGUGG,UGGUGG,UGGUGU | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| ZRANB2 | 6 | 1360 | 0.016666667 | 0.0068571141 | 1.281292 | AGGUUA,GAGGUU,GGUGGU,GUGGUG,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | CCUUCU,UCCCUU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| PABPC1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | AAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| YBX1 | 5 | 1321 | 0.014285714 | 0.0066606207 | 1.100845 | ACAUCA,AUCAUC,CAUCAU,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | GUGUGG,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF5 | 2 | 669 | 0.007142857 | 0.0033756550 | 1.081334 | GUGUGG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| G3BP1 | 14 | 3431 | 0.035714286 | 0.0172914148 | 1.046445 | ACCCCU,AGGCAG,AGGCCC,CACGCC,CAGGCC,CCAGGC,CCCAUC,CCCCCA,CCCCUC,CCCUCC,CGGCCC,UAGGCA | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.