circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000140612:-:15:84687624:84691644
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000140612:-:15:84687624:84691644 | ENSG00000140612 | MSTRG.11445.7 | - | 15 | 84687625 | 84691644 | 260 | CUCUAUUAUCAAGUCCUAAAUUUUGGAAUGAUUGUCUCAUCGGCACUAAUGAUCUGGAAGGGGUUAAUGGUAAUAACUGGAAGUGAAAGUCCGAUUGUAGUGGUGCUCAGUGGCAGCAUGGAACCUGCAUUUCAUAGAGGAGAUCUUCUCUUUCUAACAAAUCGAGUUGAAGAUCCCAUACGAGUGGGAGAAAUUGUUGUUUUUAGGAUAGAAGGAAGAGAGAUUCCUAUAGUUCACCGAGUCUUGAAGAUUCAUGAAAACUCUAUUAUCAAGUCCUAAAUUUUGGAAUGAUUGUCUCAUCGGCACUAAU | circ |
| ENSG00000140612:-:15:84687624:84691644 | ENSG00000140612 | MSTRG.11445.7 | - | 15 | 84687625 | 84691644 | 22 | AUUCAUGAAAACUCUAUUAUCA | bsj |
| ENSG00000140612:-:15:84687624:84691644 | ENSG00000140612 | MSTRG.11445.7 | - | 15 | 84691635 | 84691844 | 210 | GGAGCAGGGUGCUUUGGGUGAAAGAAGGGGGGCCUGUUUUGAUCUGCUGAUGGCAUAAUUAUUAUAAUGUAUUAAAGAAUGUCUCAGAACUGUAGUUGUCCUAAUAGUACUGCUUGAAACUUUCAAAUUGUUACUGCUUUCAGUUGCUUUCCGAAGUGGGGAUAAUGGUGGAUCUGAUCUCUUCUGUUUUGUUCUUGCAGCUCUAUUAUC | ie_up |
| ENSG00000140612:-:15:84687624:84691644 | ENSG00000140612 | MSTRG.11445.7 | - | 15 | 84687425 | 84687634 | 210 | UUCAUGAAAAGUAUGUGCAAAGUAUAGCAUCUUUGUUUCUAAGUGCUUUUGUUUAAGUGUUUGUGGUAUUUAGUUUUGAUAGCCUAAAGAUUGUAAAACAGUGAGGAACCUCUAGGCCUUCUUCCAGUCCAUUUUUAUGAAUGCUGGAAAGUUUAGUGAACAUUUUGAAAACUGUAAAUAAUUAUUUAUUAGUUCAUUUCUUUCCCUGAA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
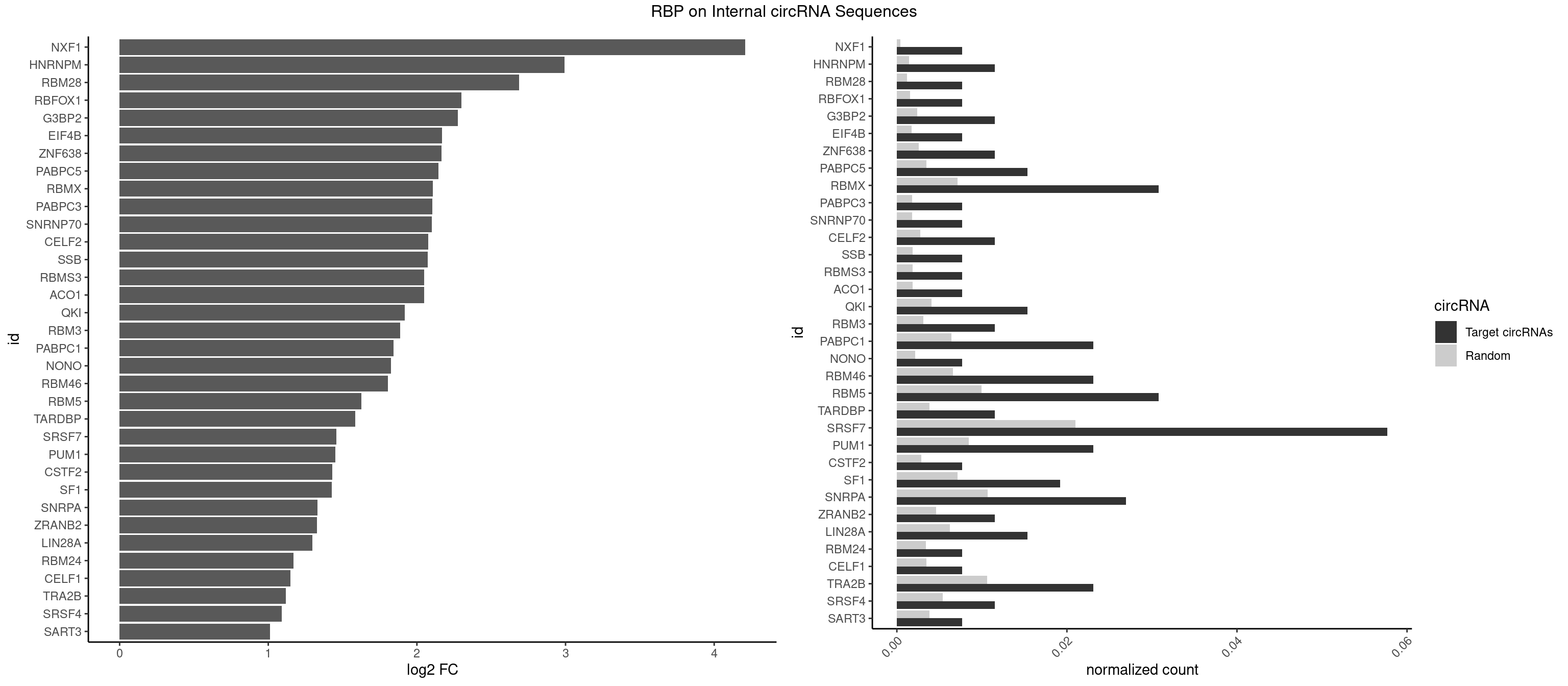
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| NXF1 | 1 | 286 | 0.007692308 | 0.0004159215 | 4.209033 | AACCUG | AACCUG |
| HNRNPM | 2 | 999 | 0.011538462 | 0.0014492040 | 2.993118 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM28 | 1 | 822 | 0.007692308 | 0.0011926949 | 2.689191 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| RBFOX1 | 1 | 1077 | 0.007692308 | 0.0015622419 | 2.299799 | AGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| G3BP2 | 2 | 1644 | 0.011538462 | 0.0023839405 | 2.275031 | AGGAUA,GGAUAG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| EIF4B | 1 | 1179 | 0.007692308 | 0.0017100607 | 2.169369 | UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ZNF638 | 2 | 1773 | 0.011538462 | 0.0025708878 | 2.166112 | GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PABPC5 | 3 | 2400 | 0.015384615 | 0.0034795387 | 2.144520 | AGAAAU,GAAAGU,GAAAUU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| RBMX | 7 | 4925 | 0.030769231 | 0.0071387787 | 2.107739 | AAGGAA,AGAAGG,AGGAAG,AUCCCA,GAAGGA,GGAAGG,UAACAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| PABPC3 | 1 | 1234 | 0.007692308 | 0.0017897669 | 2.103645 | GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| SNRNP70 | 1 | 1237 | 0.007692308 | 0.0017941145 | 2.100145 | AUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF2 | 2 | 1886 | 0.011538462 | 0.0027346479 | 2.077024 | GUUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| SSB | 1 | 1260 | 0.007692308 | 0.0018274462 | 2.073588 | UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| ACO1 | 1 | 1283 | 0.007692308 | 0.0018607779 | 2.047511 | CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMS3 | 1 | 1283 | 0.007692308 | 0.0018607779 | 2.047511 | CUAUAG | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| QKI | 3 | 2805 | 0.015384615 | 0.0040664663 | 1.919641 | ACUAAU,CACUAA,CUAACA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBM3 | 2 | 2152 | 0.011538462 | 0.0031201361 | 1.886770 | AAAACU,AUACGA | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| PABPC1 | 5 | 4443 | 0.023076923 | 0.0064402624 | 1.841260 | ACAAAU,ACUAAU,CAAAUC,CUAACA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| NONO | 1 | 1498 | 0.007692308 | 0.0021723567 | 1.824155 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM46 | 5 | 4554 | 0.023076923 | 0.0066011240 | 1.805667 | AAUGAU,AUCAAG,AUGAAA,AUGAUU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| RBM5 | 7 | 6879 | 0.030769231 | 0.0099705232 | 1.625747 | AAGGAA,AAGGGG,CUUCUC,GAAGGA,GAAGGG,UCUUCU,UUCUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| TARDBP | 2 | 2654 | 0.011538462 | 0.0038476365 | 1.584406 | GAAUGA,GUUGUU | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| SRSF7 | 14 | 14481 | 0.057692308 | 0.0209873716 | 1.458857 | AAUGAU,AGAGAG,AGAGAU,AGAGGA,AGAUCU,AGGAAG,AUAGAA,CGAUUG,GAAUGA,GAGAGA,GAGAUC,GAUAGA,GGAAGG | AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| PUM1 | 5 | 5823 | 0.023076923 | 0.0084401638 | 1.451108 | AAUUGU,AUUGUA,GUAAUA,UUAAUG,UUGUAG | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| CSTF2 | 1 | 1967 | 0.007692308 | 0.0028520334 | 1.431426 | UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| SF1 | 4 | 4931 | 0.019230769 | 0.0071474739 | 1.427911 | ACUAAU,CACCGA,CUAACA,UAACAA | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| SNRPA | 6 | 7380 | 0.026923077 | 0.0106965744 | 1.331694 | ACCUGC,AGGAGA,AUUCCU,GAUUCC,GGAGAU,GUAGUG | ACCUGC,AGCAGU,AGGAGA,AGUAGG,AGUAGU,AUACCU,AUGCAC,AUGCUG,AUUCCU,AUUGCA,CAGUAG,CCUGCU,CUGCUA,GAGCAG,GAUACC,GAUUCC,GCAGUA,GGAGAU,GGGUAU,GGUAUG,GUAGGC,GUAGGG,GUAGUC,GUAGUG,GUAUGC,GUUACC,GUUUCC,UACCUG,UAUGCU,UCCUGC,UGCACA,UGCACC,UGCACG,UUACCU,UUCCUG,UUGCAC,UUUCCU |
| ZRANB2 | 2 | 3173 | 0.011538462 | 0.0045997733 | 1.326816 | GUGGUG,UGGUAA | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| LIN28A | 3 | 4315 | 0.015384615 | 0.0062547643 | 1.298461 | AGGAGA,GGAGAA,GGAGAU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| RBM24 | 1 | 2357 | 0.007692308 | 0.0034172229 | 1.170592 | GAGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| CELF1 | 1 | 2391 | 0.007692308 | 0.0034664959 | 1.149938 | UGUUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| TRA2B | 5 | 7329 | 0.023076923 | 0.0106226650 | 1.119305 | AAGGAA,AGAAGG,AGGAAG,GAAGGA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SRSF4 | 2 | 3740 | 0.011538462 | 0.0054214720 | 1.089694 | AGGAAG,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SART3 | 1 | 2634 | 0.007692308 | 0.0038186524 | 1.010353 | GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
RBP on BSJ (Exon Only)
Plot
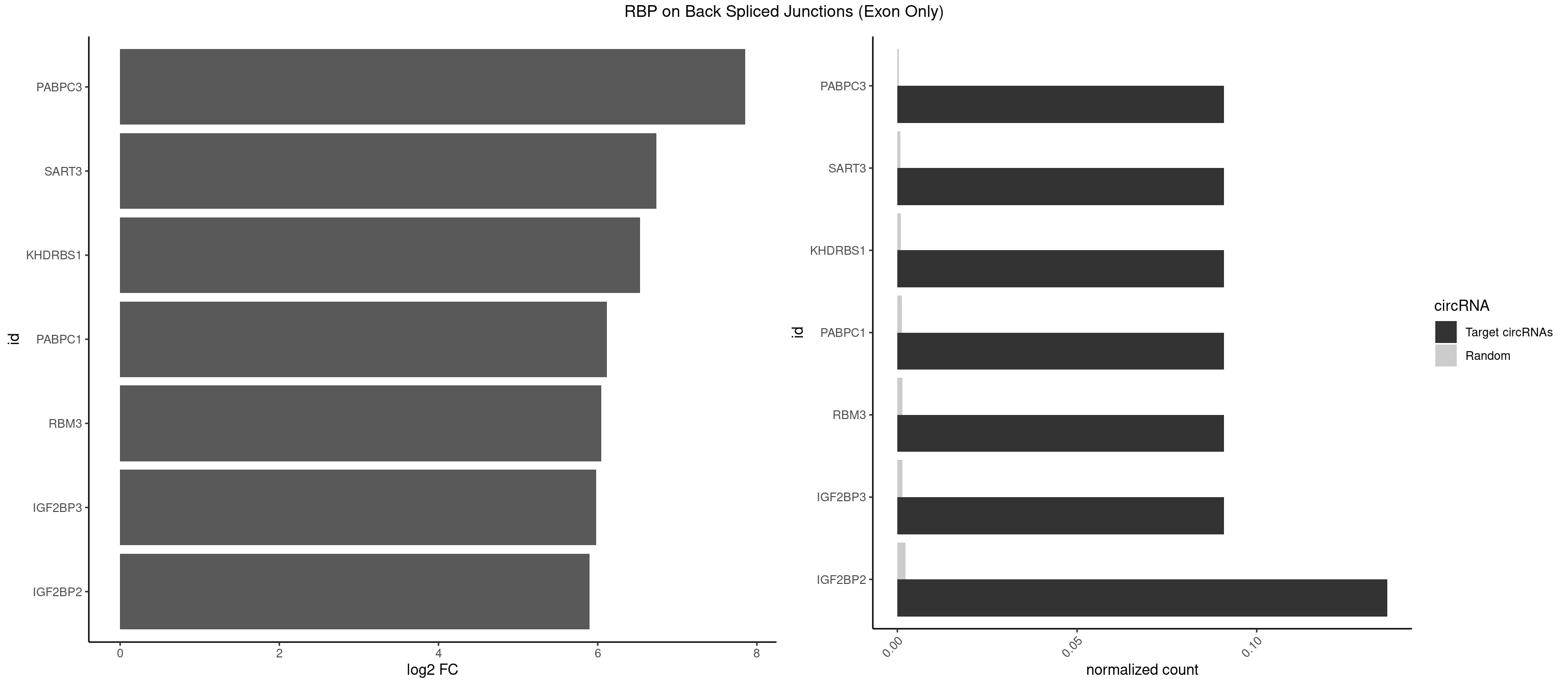
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| PABPC3 | 1 | 5 | 0.09090909 | 0.0003929788 | 7.853829 | GAAAAC | AAAACC,GAAAAC |
| SART3 | 1 | 12 | 0.09090909 | 0.0008514540 | 6.738352 | GAAAAC | AAAAAA,AGAAAA,GAAAAA,GAAAAC |
| KHDRBS1 | 1 | 14 | 0.09090909 | 0.0009824469 | 6.531901 | GAAAAC | AUUUAA,CUAAAA,GAAAAC,UAAAAA,UAAAAG,UUAAAA,UUAAAU |
| PABPC1 | 1 | 19 | 0.09090909 | 0.0013099293 | 6.116864 | GAAAAC | AAAAAA,ACAAAC,ACGAAU,AGAAAA,CAAACC,CAAAUA,CGAACA,CUAAUA,GAAAAA,GAAAAC |
| RBM3 | 1 | 20 | 0.09090909 | 0.0013754257 | 6.046474 | AAAACU | AAAACG,AAAACU,AAGACU,AAUACG,AAUACU,AUACUA,GAAACU,GAGACG,GAGACU,GAUACU |
| IGF2BP3 | 1 | 21 | 0.09090909 | 0.0014409222 | 5.979360 | AAACUC | AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,ACAAAC,ACAAUC,ACACUC,ACAUAC,CAAUCA,CACUCA,CAUACA,CAUUCA |
| IGF2BP2 | 2 | 34 | 0.13636364 | 0.0022923762 | 5.894471 | AAACUC,GAAAAC | AAAAUC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACUCA,ACAAAC,ACAAUC,ACACUC,ACAUAC,CAAAAC,CAAUAC,CAAUCA,CAAUUC,CACUCA,CAUACA,CAUUCA,CCAUAC,CCAUUC,GAAAAC,GAACAC,GAAUAC,GAAUUC |
RBP on BSJ (Exon and Intron)
Plot
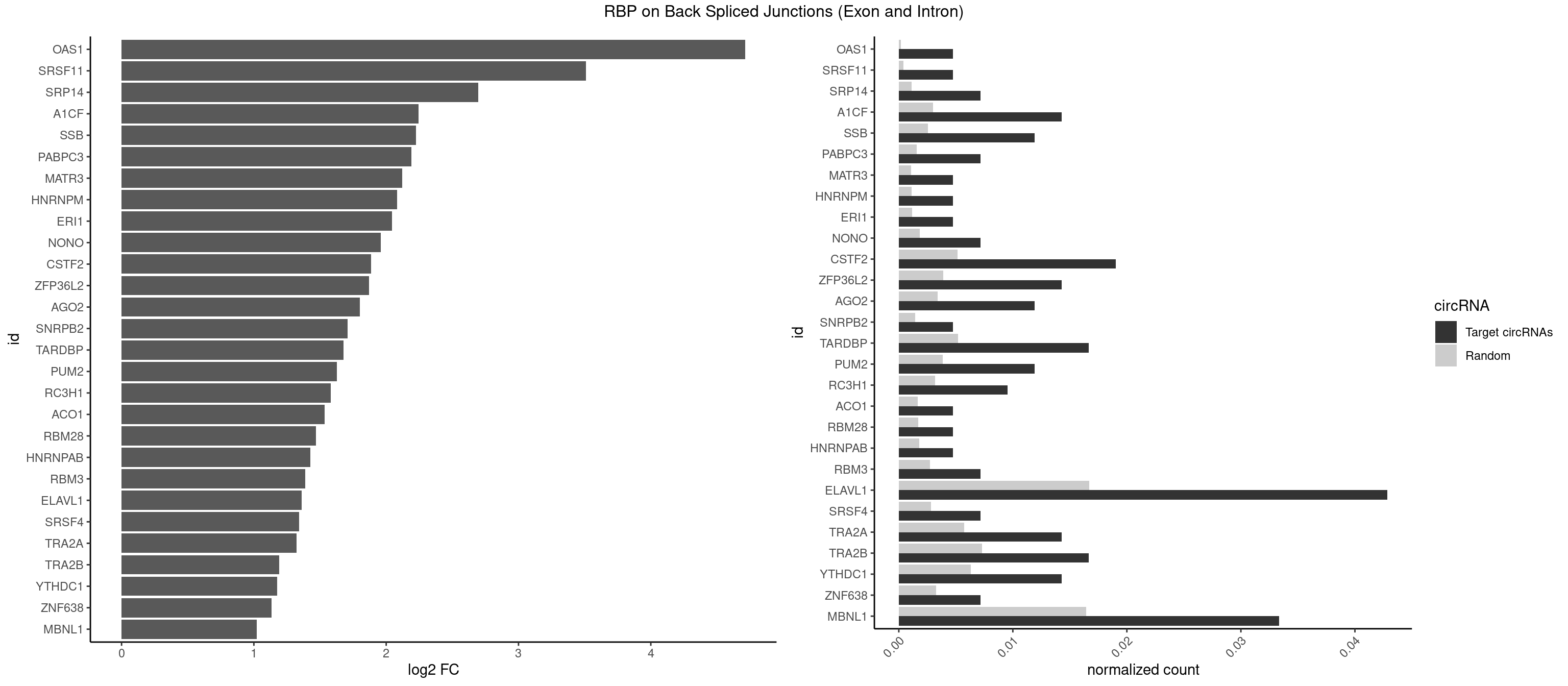
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 35 | 0.004761905 | 0.0001813785 | 4.714464 | GCAUAA | GCAUAA |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CUGUAG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| A1CF | 5 | 598 | 0.014285714 | 0.0030179363 | 2.242939 | AGUAUA,AUAAUU,UAAUUA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| SSB | 4 | 505 | 0.011904762 | 0.0025493753 | 2.223323 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| HNRNPM | 1 | 222 | 0.004761905 | 0.0011235389 | 2.083489 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| ERI1 | 1 | 228 | 0.004761905 | 0.0011537686 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| NONO | 2 | 364 | 0.007142857 | 0.0018389762 | 1.957598 | AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| CSTF2 | 7 | 1022 | 0.019047619 | 0.0051541717 | 1.885798 | GUGUUU,GUUUUG,UGUUUG,UGUUUU | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| ZFP36L2 | 5 | 774 | 0.014285714 | 0.0039046755 | 1.871299 | AUUUAU,UAUUUA,UUAUUU,UUUAUU | AUUUAU,UAUUUA,UUAUUU,UUUAUU |
| AGO2 | 4 | 677 | 0.011904762 | 0.0034159613 | 1.801175 | AAGUGC,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| TARDBP | 6 | 1034 | 0.016666667 | 0.0052146312 | 1.676328 | GAAUGU,GUUUUG,UGAAUG,UUGUUC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| PUM2 | 4 | 764 | 0.011904762 | 0.0038542926 | 1.627001 | GUAAAU,UAAAUA,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CCUUCU,CUUCUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGU | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM3 | 2 | 541 | 0.007142857 | 0.0027307537 | 1.387202 | AAAACU,GAAACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| ELAVL1 | 17 | 3309 | 0.042857143 | 0.0166767432 | 1.361698 | AUUUAU,UAGUUU,UAUUUA,UGUUUU,UUAGUU,UUAUUU,UUGUUU,UUUAGU,UUUAUU,UUUGUU | AUUUAU,CCCCCC,UAGUUU,UAUUUA,UAUUUU,UGAUUU,UGGUUU,UGUUUU,UUAGUU,UUAUUU,UUGAUU,UUGGUU,UUGUUU,UUUAGU,UUUAUU,UUUGGU,UUUGUU,UUUUUG,UUUUUU |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AAGAAG,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| TRA2A | 5 | 1133 | 0.014285714 | 0.0057134220 | 1.322146 | AAAGAA,AAGAAG,GAAAGA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AAAGAA,AAGAAG,AAGAAU,AGAAGG,GAAAGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| YTHDC1 | 5 | 1254 | 0.014285714 | 0.0063230552 | 1.175879 | GAAUGC,GGGUGC,UACUGC,UAGUAC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GUUCUU,UGUUCU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| MBNL1 | 13 | 3258 | 0.033333333 | 0.0164197904 | 1.021530 | CUGCUG,CUGCUU,GCUUUU,GUGCUU,UCUGCU,UGCUUU,UGUCUC,UUGCUU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.