circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000196739:+:9:114162714:114183075
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000196739:+:9:114162714:114183075 | ENSG00000196739 | ENST00000356083 | + | 9 | 114162715 | 114183075 | 1954 | GGGGUUUCUCUUCUCCUGGAUCUUAGUCUCGUUUGCCUGUCACCUGGCCUCCACCCAAGGAGCUCCUGAAGAUGUGGACAUCCUCCAGCGGCUGGGCCUCAGCUGGACGAAGGCCGGGAGCCCUGCACCCCCGGGAGUCAUUCCUUUCCAGUCGGGCUUCAUCUUUACGCAGCGGGCCCGGCUCCAGGCUCCCACGGGCACCGUCAUUCCUGCCGCCUUGGGCACAGAGCUGGCACUGGUGCUGAGCCUCUGCUCCCACCGGGUGAACCAUGCCUUCCUCUUCGCUGUCCGCAGCCAGAAACGCAAGCUGCAGCUGGGCCUGCAGUUCCUCCCCGGCAAGACGGUCGUCCACCUCGGGUCCCGGCGCUCAGUGGCCUUCGACCUCGACAUGCACGACGGGCGCUGGCACCACCUGGCCCUCGAGCUCCGAGGCCGCACAGUCACUCUGGUGACUGCCUGCGGGCAGCGCCGGGUGCCUGUCCUGCUGCCUUUCCACAGGGACCCUGCACUCGACCCUGGGGGCUCCUUCCUCUUUGGGAAGAUGAACCCGCAUGCAGUCCAGUUUGAAGGUGCUCUCUGCCAGUUCAGUAUCUACCCUGUGACGCAGGUCGCUCACAAUUACUGUACCCACCUGAGGAAGCAGUGUGGACAGGCUGACACGUACCAGUCCCCACUGGGACCUCUCUUCUCCCAAGACUCUGGCAGACCUUUUACCUUCCAGUCCGACCUCGCCCUGCUAGGCCUGGAGAACUUGACCACUGCCACACCAGCCCUGGGGUCACUGCCAGCAGGCAGGGGACCCAGGGGGACUGUGGCACCCGCCACGCCCACCAAGCCCCAAAGGACUAGCCCCACAAACCCUCACCAGCAUAUGGCGGUGGGAGGCCCAGCCCAAACCCCGCUGCUACCUGCCAAGCUGUCAGCCAGUAACGCACUUGAUCCCAUGCUCCCAGCCUCUGUUGGCGGCUCUACCAGAACGCCUCGCCCUGCGGCCGCUCAACCAUCACAGAAGAUCACAGCCACCAAAAUCCCCAAAAGCCUCCCUACCAAGCCUUCGGCCCCUUCUACUUCAAUUGUGCCCAUCAAAAGCCCCCAUCCUACCCAGAAAACAGCUCCAUCUUCAUUUACAAAGUCAGCCCUACCCACUCAGAAGCAAGUGCCACCUACUUCCCGUCCAGUUCCUGCCAGAGUCUCCCGUCCCGCAGAGAAGCCCAUCCAGAGGAACCCGGGAAUGCCCAGGCCCCCACCGCCCAGCACCCGGCCCCUACCUCCUACCACCAGCUCCUCUAAAAAACCCAUUCCCACACUAGCUCGGACUGAGGCCAAGAUAACCAGCCAUGCCAGUAAGCCGGCCUCUGCCCGCACCAGCACCCACAAACCUCCCCCAUUUACUGCUUUAUCCUCAUCUCCUGCCCCUACUCCUGGUUCUACCAGGAGUACUCGGCCACCAGCCACGAUGGUACCUCCAACUUCGGGCACCAGCACUCCCAGAACAGCACCUGCCGUCCCCACUCCUGGCUCAGCUCCCACUGGAAGCAAGAAGCCCAUUGGAUCGGAAGCCUCAAAGAAAGCCGGACCCAAGAGCAGCCCCCGGAAGCCUGUCCCCCUCAGACCUGGGAAGGCAGCCAGGGAUGUCCCCUUGAGCGAUCUGACAACCAGGCCUAGCCCCAGACAGCCCCAGCCCAGUCAGCAGACCACCCCGGCCCUGGUAUUGGCCCCGGCGCAAUUCCUGUCCUCCAGCCCCCGGCCCACGAGCAGUGGCUAUUCGAUCUUCCACCUGGCAGGAUCUACGCCUUUCCCUCUGCUGAUGGGGCCUCCGGGACCCAAGGGAGACUGUGGCUUGCCGGGUCCCCCUGGGCUACCUGGGCUACCUGGAAUCCCUGGUGCACGUGGGCCUCGGGGUCCUCCUGGGCCUUAUGGAAAUCCAGGUCUCCCCGGCCCUCCUGGAGCCAAAGGGGUUUCUCUUCUCCUGGAUCUUAGUCUCGUUUGCCUGUCACCUGGCCU | circ |
| ENSG00000196739:+:9:114162714:114183075 | ENSG00000196739 | ENST00000356083 | + | 9 | 114162715 | 114183075 | 22 | CUGGAGCCAAAGGGGUUUCUCU | bsj |
| ENSG00000196739:+:9:114162714:114183075 | ENSG00000196739 | ENST00000356083 | + | 9 | 114162515 | 114162724 | 210 | ACUGAGAGGCUGUGCCUGGAGGCCGGGAAGAGUGGGCUGCUGCUCUGAUGGGGUCCUAGAGCCUGUACCUGGCCUCCUGCCUCCCGUGGGCAGGACUGGGGUGAGACUGGGACUGGUUUCCACUCCCAGCUUCCCCAGCCGGAUCCGGGUGUGAGGGGUGGAAGCUGAUGCCGCCUCUGCUUGUGUCUCCUGGUCUCUAGGGGGUUUCUC | ie_up |
| ENSG00000196739:+:9:114162714:114183075 | ENSG00000196739 | ENST00000356083 | + | 9 | 114183066 | 114183275 | 210 | UGGAGCCAAAGUGAGUAUUUGCUGGAGAUGUGGCCAUGGAGUGGGUGCUGGGGUUGGAGCCAUGUUUGCAGUGCUUCCCAGGGGUGCCUGGUGGGAGGGCUUCAGGGGAACUGAGGGGGAUUCCCGCCUAAGCCAGGGGCACCCUCUGGGCCCCUUUCCAGGGCUGGGCCCUGUGCCUGGUCCGGAGACACAGUGCUUCCGUCUGGGAGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
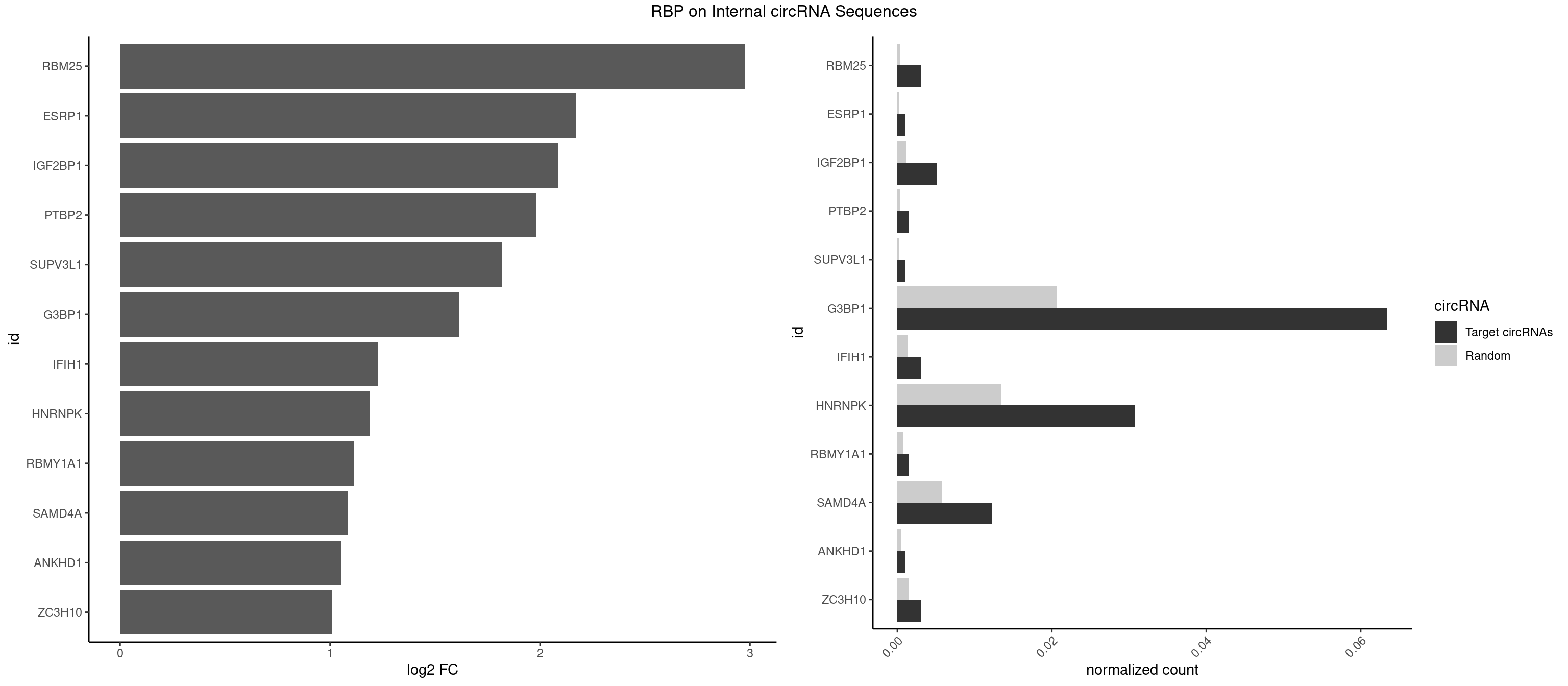
RBP on full sequence
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 5 | 268 | 0.003070624 | 0.0003898359 | 2.977593 | CGGGCA,UCGGGC | AUCGGG,CGGGCA,UCGGGC |
| ESRP1 | 1 | 156 | 0.001023541 | 0.0002275250 | 2.169472 | AGGGAU | AGGGAU |
| IGF2BP1 | 9 | 831 | 0.005117707 | 0.0012057377 | 2.085582 | AGCACC,CACCCG,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PTBP2 | 2 | 267 | 0.001535312 | 0.0003883867 | 1.982966 | CUCUCU | CUCUCU |
| SUPV3L1 | 1 | 199 | 0.001023541 | 0.0002898408 | 1.820237 | CCGCCC | CCGCCC |
| G3BP1 | 123 | 14282 | 0.063459570 | 0.0206989801 | 1.616278 | ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCGC,ACGCAC,ACGCAG,ACGCCC,AGGCAG,AGGCCC,AGGCCG,CACAGG,CACCCG,CACCGG,CACGCC,CAGGCA,CAGGCC,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUCC,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGGCA,CCGGCC,CCUCCG,CCUCGG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCGGC,UACGCA,UACGCC,UAGGCC,UCCGCA,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| IFIH1 | 5 | 904 | 0.003070624 | 0.0013115296 | 1.227282 | GGCCCU,GGCCGC | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| HNRNPK | 59 | 9304 | 0.030706244 | 0.0134848428 | 1.187193 | AAAAAA,ACCCAA,ACCCAU,ACCCCC,ACCCGC,CAAACC,CCAAAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCUA | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| RBMY1A1 | 2 | 489 | 0.001535312 | 0.0007101099 | 1.112418 | CAAGAC | ACAAGA,CAAGAC |
| SAMD4A | 23 | 3992 | 0.012282497 | 0.0057866714 | 1.085798 | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,GCGGGC,GCUGGA,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| ANKHD1 | 1 | 339 | 0.001023541 | 0.0004927293 | 1.054702 | GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| ZC3H10 | 5 | 1053 | 0.003070624 | 0.0015274610 | 1.007397 | CAGCGC,CCAGCG,GAGCGA,GCAGCG | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
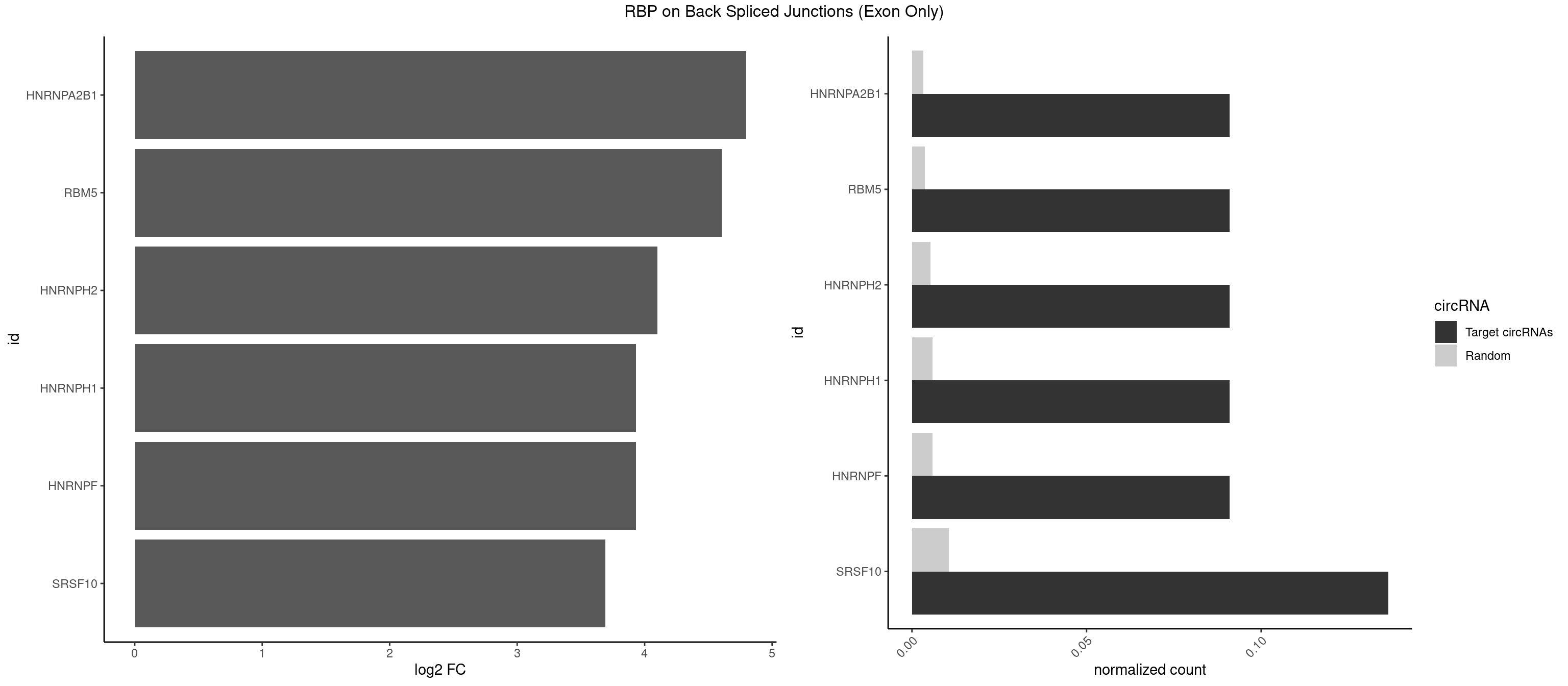
RBP on BSJ (Exon Only)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| HNRNPA2B1 | 1 | 49 | 0.09090909 | 0.003274823 | 4.794936 | AAGGGG | AAGGAA,AAGGGG,AAUUUA,AGAAGC,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,GAAGCC,GAAGGA,GCCAAG,GCGAAG,GGAACC,GGGGCC,UAGACA,UUAGGG |
| RBM5 | 1 | 56 | 0.09090909 | 0.003733298 | 4.605902 | AAGGGG | AAAAAA,AAGGAA,AAGGGG,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CUCUUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGUGGU,UCUUCU |
| HNRNPH2 | 1 | 80 | 0.09090909 | 0.005305214 | 4.098942 | AAGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| HNRNPF | 1 | 90 | 0.09090909 | 0.005960178 | 3.930997 | AAGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,CGAUGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH1 | 1 | 90 | 0.09090909 | 0.005960178 | 3.930997 | AAGGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AGGAAG,AGGGAA,AGGGGA,AUUGGG,CAGGAC,CGAGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGUGU,UCGGGC,UGGGGU,UGGGUG,UGUGGG |
| SRSF10 | 2 | 160 | 0.13636364 | 0.010544931 | 3.692837 | AAAGGG,AAGGGG | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGG,AAGAAA,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
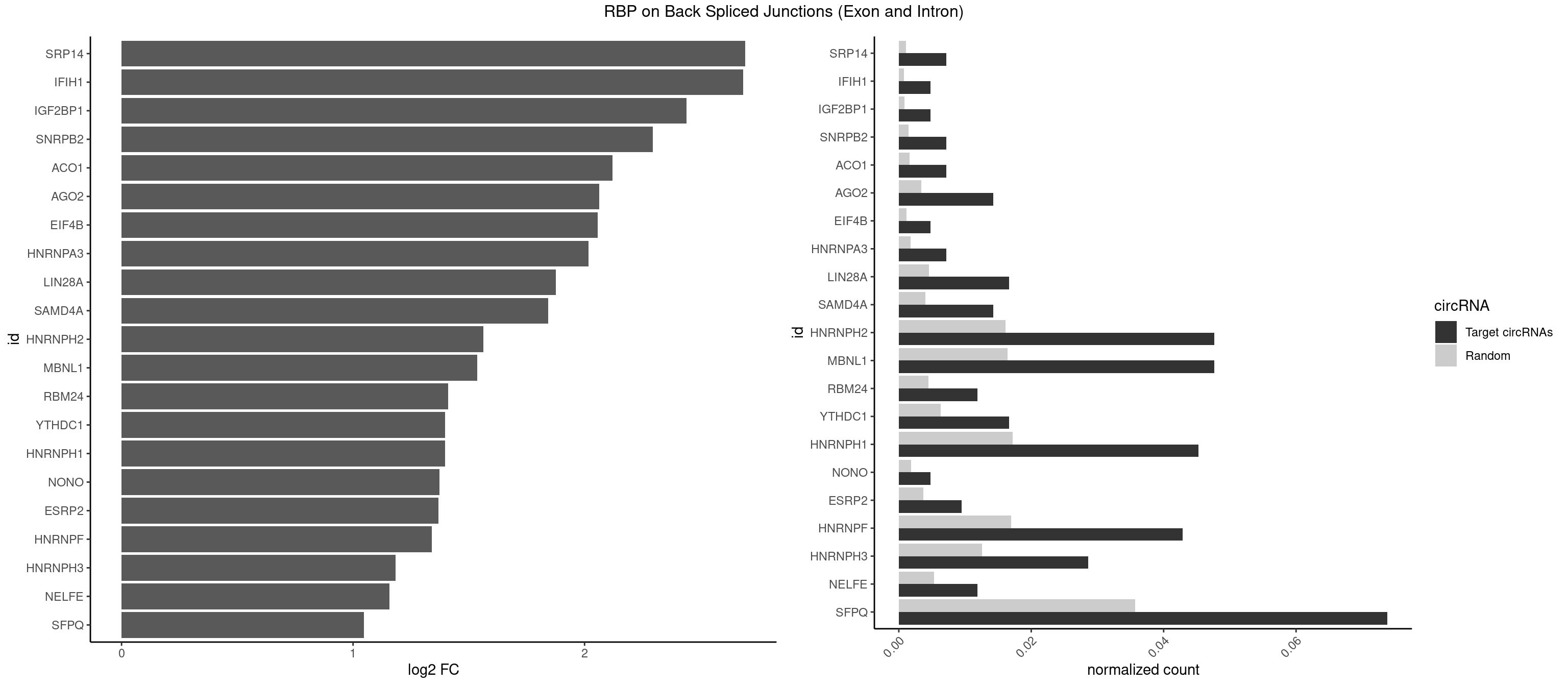
RBP on BSJ (Exon and Intron)
Plot
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRP14 | 2 | 218 | 0.007142857 | 0.0011033857 | 2.694564 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UGCAGU,UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| AGO2 | 5 | 677 | 0.014285714 | 0.0034159613 | 2.064210 | AAAGUG,AGUGCU,GUGCUU | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| EIF4B | 1 | 226 | 0.004761905 | 0.0011436921 | 2.057840 | GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| LIN28A | 6 | 900 | 0.016666667 | 0.0045395002 | 1.876360 | CGGAGA,GGAGAU,GGAGGG,UGGAGA,UGGAGG,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SAMD4A | 5 | 790 | 0.014285714 | 0.0039852882 | 1.841817 | CGGGAA,CUGGCC,CUGGUC,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| HNRNPH2 | 19 | 3198 | 0.047619048 | 0.0161174929 | 1.562911 | AGGGGA,AUGUGG,CAGGAC,CUGGGG,GAGGGG,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAA,GGGGGA,GGGUGU,UGGGGU,UGGGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| MBNL1 | 19 | 3258 | 0.047619048 | 0.0164197904 | 1.536103 | AUGCCG,CUGCCU,CUGCUC,CUGCUG,CUGCUU,CUUGUG,GCUGCU,GCUUGU,GUGCUG,GUGCUU,UCUGCU,UGCUGC,UGCUUC,UGUCUC,UUGCUG,UUUGCU | ACGCUA,ACGCUC,ACGCUG,ACGCUU,AUGCCA,AUGCCC,AUGCCG,AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUC,CCGCUG,CCGCUU,CCUGCU,CGCUGC,CGCUGU,CGCUUC,CGCUUG,CGCUUU,CUCGCU,CUGCCA,CUGCCC,CUGCCG,CUGCCU,CUGCGG,CUGCUA,CUGCUC,CUGCUG,CUGCUU,CUUGCU,CUUGUG,GCGCUC,GCGCUG,GCGCUU,GCUGCG,GCUGCU,GCUUGC,GCUUGU,GCUUUU,GGCUUU,GUCUCG,GUGCUA,GUGCUC,GUGCUG,GUGCUU,UCCGCU,UCGCUA,UCGCUC,UCGCUG,UCGCUU,UCUCGC,UCUGCU,UGCGGC,UGCUGC,UGCUGU,UGCUUC,UGCUUU,UGUCUC,UUCGCU,UUGCCA,UUGCCC,UUGCCG,UUGCCU,UUGCUA,UUGCUC,UUGCUG,UUGCUU,UUGUGC,UUUGCU |
| RBM24 | 4 | 888 | 0.011904762 | 0.0044790407 | 1.410277 | AGAGUG,GAGUGG,GUGUGA | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| YTHDC1 | 6 | 1254 | 0.016666667 | 0.0063230552 | 1.398272 | GGCUGC,GGGUGC,UCCUGC,UGAUGC,UGCUGC | GAAUAC,GAAUGC,GACUAC,GACUGC,GAGUAC,GAGUGC,GCAUAC,GCAUGC,GCCUAC,GCCUGC,GCGUAC,GCGUGC,GGAUAC,GGAUGC,GGCUAC,GGCUGC,GGGUAC,GGGUGC,UAAUAC,UAAUGC,UACUAC,UACUGC,UAGUAC,UAGUGC,UCAUAC,UCAUGC,UCCUAC,UCCUGC,UCGUAC,UCGUGC,UGAUAC,UGAUGC,UGCUAC,UGCUGC,UGGUAC,UGGUGC |
| HNRNPH1 | 18 | 3406 | 0.045238095 | 0.0171654575 | 1.398030 | AGGGGA,AUGUGG,CAGGAC,CUGGGG,GAGGGG,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAA,GGGUGU,UGGGGU,UGGGUG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGAGG | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAG,GGGGAA,GGGGAU | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPF | 17 | 3360 | 0.042857143 | 0.0169336961 | 1.339639 | AGGGGA,AUGGGG,AUGUGG,CUGGGG,GAGGGG,GAUGGG,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGGAA,UGGGGU | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| HNRNPH3 | 11 | 2496 | 0.028571429 | 0.0125806127 | 1.183371 | AGGGGA,AUGUGG,GAGGGG,GGAGGG,GGGAAG,GGGAGG,GGGCUG,GGGUGU,UGGGUG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| NELFE | 4 | 1059 | 0.011904762 | 0.0053405885 | 1.156468 | CUCUGG,CUGGUU,GGUCUC,GUCUCU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| SFPQ | 30 | 7087 | 0.073809524 | 0.0357114067 | 1.047422 | ACUGGG,AGGGGG,CUGGAG,CUGGGA,GAAGAG,GACUGG,GAGAGG,GGAAGA,GGACUG,GGAGGG,GGGGAU,GGGGGA,GUCUGG,UAGGGG,UGAUGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUUU | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.