circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000138814:-:4:101195915:101196116
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000138814:-:4:101195915:101196116 | ENSG00000138814 | ENST00000394854 | - | 4 | 101195916 | 101196116 | 201 | CUGUUCCAUUUCCUCCAAGUCACCGGCUUACAGCAAAAGAAGUGUUUGAUAAUGAUGGAAAACCUCGUGUGGAUAUCUUAAAGGCGCAUCUUAUGAAGGAGGGAAGGCUGGAAGAGAGUGUUGCAUUGAGAAUAAUAACAGAGGGUGCAUCAAUUCUUCGACAGGAAAAAAAUUUGCUGGAUAUUGAUGCGCCAGUCACUGCUGUUCCAUUUCCUCCAAGUCACCGGCUUACAGCAAAAGAAGUGUUUGAU | circ |
| ENSG00000138814:-:4:101195915:101196116 | ENSG00000138814 | ENST00000394854 | - | 4 | 101195916 | 101196116 | 22 | GCCAGUCACUGCUGUUCCAUUU | bsj |
| ENSG00000138814:-:4:101195915:101196116 | ENSG00000138814 | ENST00000394854 | - | 4 | 101196107 | 101196316 | 210 | GCAAUCACAUUAUAUCUAGAAUCAAAGGAAGGAUGCAGUUCUUAAUUUUUGAUAGAUAGCCCUUGAUCAUAUUUUAAUUCAGCCACUCUAAAAAUAUUAUAUUAGUUGGUUUGUGAUGAGGAUGGAUAUUUGGUGGUUAUUAUUGCAUUGUUUGUUGUUGAGUUUUGGCUAAUGUAAUAAUUAGACCUUUCUUCUUUCAGCUGUUCCAUU | ie_up |
| ENSG00000138814:-:4:101195915:101196116 | ENSG00000138814 | ENST00000394854 | - | 4 | 101195716 | 101195925 | 210 | CCAGUCACUGGUAAGUCCUGAGGAGGGAGGUUGCCCACUUGUAUACACAUUUUGUUGUUUAUAGCAUGAAAUCAAAAGUCCUAGUUACCAGUGAGUCUUCAAAUACCUUACCCAUCAUAAUCUUUUACUGUUGAAUCUAUACAUUAGAGUAGUAAGUAUAUUAUACAUAUAUACACAUAUUUUCAUGUGUUAAAUAUCUGUAAGAUUGCA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
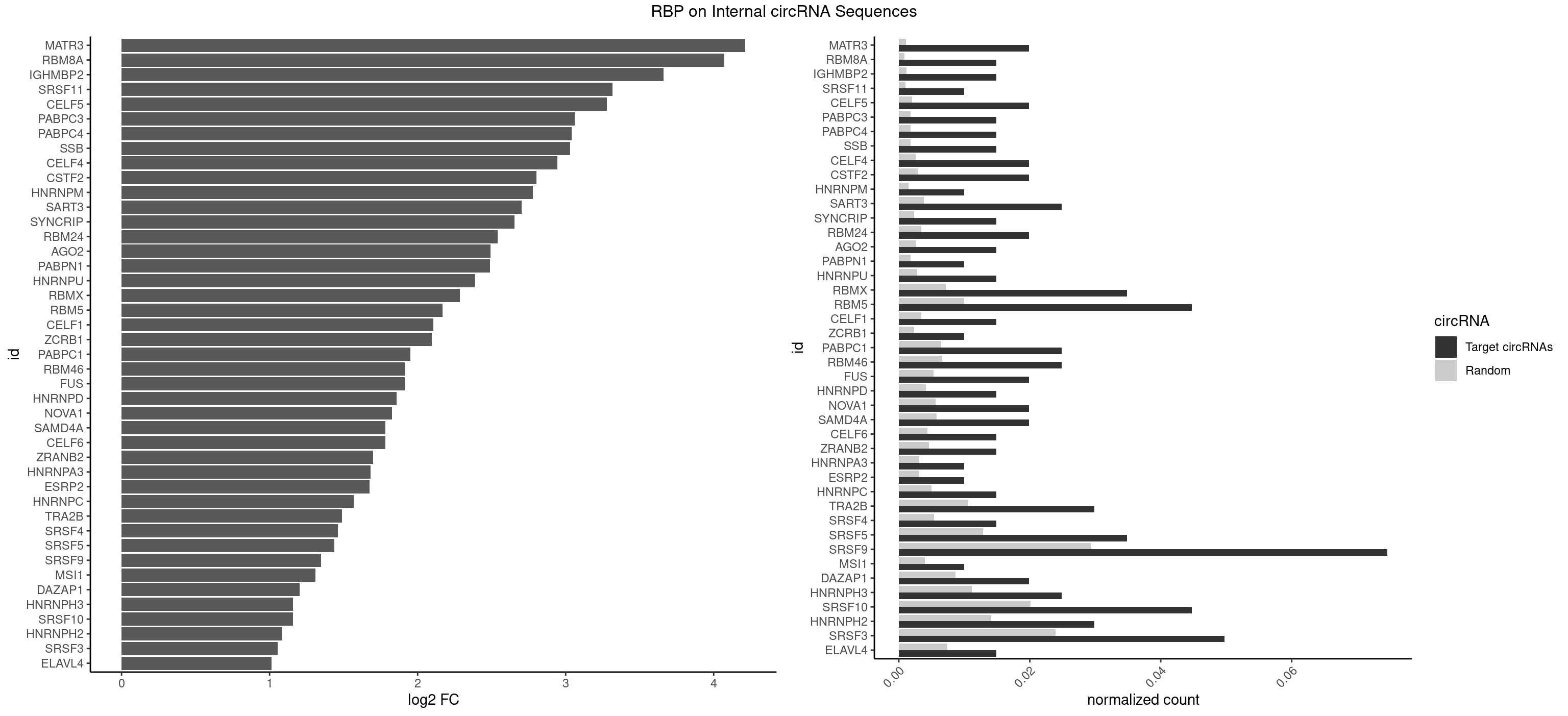
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| MATR3 | 3 | 739 | 0.019900498 | 0.0010724109 | 4.213875 | AUCUUA,CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM8A | 2 | 611 | 0.014925373 | 0.0008869128 | 4.072831 | AUGCGC,UGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| IGHMBP2 | 2 | 813 | 0.014925373 | 0.0011796520 | 3.661334 | AAAAAA | AAAAAA |
| SRSF11 | 1 | 688 | 0.009950249 | 0.0009985015 | 3.316896 | AAGAAG | AAGAAG |
| CELF5 | 3 | 1415 | 0.019900498 | 0.0020520728 | 3.277651 | GUGUGG,GUGUUG,GUGUUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| PABPC3 | 2 | 1234 | 0.014925373 | 0.0017897669 | 3.059923 | AAAACC,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| PABPC4 | 2 | 1251 | 0.014925373 | 0.0018144033 | 3.040200 | AAAAAA | AAAAAA,AAAAAG |
| SSB | 2 | 1260 | 0.014925373 | 0.0018274462 | 3.029866 | GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| CELF4 | 3 | 1782 | 0.019900498 | 0.0025839306 | 2.945165 | GUGUGG,GUGUUG,GUGUUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CSTF2 | 3 | 1967 | 0.019900498 | 0.0028520334 | 2.802742 | GUGUUG,GUGUUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| HNRNPM | 1 | 999 | 0.009950249 | 0.0014492040 | 2.779472 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| SART3 | 4 | 2634 | 0.024875622 | 0.0038186524 | 2.703597 | AAAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| SYNCRIP | 2 | 1634 | 0.014925373 | 0.0023694485 | 2.655144 | AAAAAA | AAAAAA,UUUUUU |
| RBM24 | 3 | 2357 | 0.019900498 | 0.0034172229 | 2.541908 | AGAGUG,GAGUGU,GUGUGG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| AGO2 | 2 | 1830 | 0.014925373 | 0.0026534924 | 2.491803 | AAAAAA | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PABPN1 | 1 | 1222 | 0.009950249 | 0.0017723764 | 2.489048 | AAAAGA | AAAAGA,AGAAGA |
| HNRNPU | 2 | 1965 | 0.014925373 | 0.0028491350 | 2.389171 | AAAAAA | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| RBMX | 6 | 4925 | 0.034825871 | 0.0071387787 | 2.286410 | AAGAAG,AAGUGU,AGUGUU,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| RBM5 | 8 | 6879 | 0.044776119 | 0.0099705232 | 2.166988 | AAAAAA,AAGGAG,AGGGAA,AGGGUG,GAAGGA,GAGGGA,GAGGGU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| CELF1 | 2 | 2391 | 0.014925373 | 0.0034664959 | 2.106217 | GUGUUG,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ZCRB1 | 1 | 1605 | 0.009950249 | 0.0023274215 | 2.096000 | GGCUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PABPC1 | 4 | 4443 | 0.024875622 | 0.0064402624 | 1.949541 | AAAAAA,GAAAAA,GAAAAC | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| RBM46 | 4 | 4554 | 0.024875622 | 0.0066011240 | 1.913949 | AAUGAU,AUCAAU,AUGAAG,AUGAUG | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| FUS | 3 | 3648 | 0.019900498 | 0.0052881452 | 1.911971 | AAAAAA,GGGUGC | AAAAAA,CGGUGA,CGGUGG,GGGGGG,GGGUGA,GGGUGC,GGGUGG,GGGUGU,UGGUGA,UGGUGG,UGGUGU,UUUUUU |
| HNRNPD | 2 | 2837 | 0.014925373 | 0.0041128408 | 1.859560 | AAAAAA | AAAAAA,AAUUUA,AGAUAU,AGUAGG,AUUUAU,UAUUUA,UUAGAG,UUAGGA,UUAGGG,UUAUUU,UUUAUU |
| NOVA1 | 3 | 3872 | 0.019900498 | 0.0056127669 | 1.826020 | AGUCAC,CAGUCA | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| SAMD4A | 3 | 3992 | 0.019900498 | 0.0057866714 | 1.781999 | CUGGAA,GCUGGA | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| CELF6 | 2 | 2997 | 0.014925373 | 0.0043447134 | 1.780434 | GUGUGG,GUGUUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| ZRANB2 | 2 | 3173 | 0.014925373 | 0.0045997733 | 1.698132 | AGGGAA,UAAAGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| HNRNPA3 | 1 | 2140 | 0.009950249 | 0.0031027457 | 1.681187 | AAGGAG | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| ESRP2 | 1 | 2150 | 0.009950249 | 0.0031172377 | 1.674464 | GGGAAG | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPC | 2 | 3471 | 0.014925373 | 0.0050316361 | 1.568668 | GGAUAU | AUUUUU,CUUUUU,GCAUAC,GGAUAC,GGAUAU,GGAUUC,GGGGGG,GGGUAC,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
| TRA2B | 5 | 7329 | 0.029850746 | 0.0106226650 | 1.490621 | AAAGAA,AAGAAG,AGGAAA,GAAGGA,GGAAGG | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| SRSF4 | 2 | 3740 | 0.014925373 | 0.0054214720 | 1.461010 | AAGAAG,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SRSF5 | 6 | 8869 | 0.034825871 | 0.0128544391 | 1.437893 | AAGAAG,CGCAUC,GGAAGA,UAAAGG,UACAGC,UGCAUC | AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| SRSF9 | 14 | 20254 | 0.074626866 | 0.0293536261 | 1.346156 | AGGAAA,CUGGAU,GAAGGA,GAAGGC,GAUGGA,GGAAAA,GGAAGG,GGGUGC,GGUGCA,UGAAGG,UGAGAA,UGAUGC,UGAUGG | AGGAAA,AGGAAC,AGGACA,AGGACC,AGGAGA,AGGAGC,AUGAAA,AUGAAC,AUGACA,AUGACC,AUGAGA,AUGAGC,CUGGAU,GAAGCA,GAAGCC,GAAGGA,GAAGGC,GAUGCA,GAUGCC,GAUGGA,GAUGGC,GGAAAA,GGAAAG,GGAACA,GGAACG,GGAAGC,GGAAGG,GGACAA,GGACAG,GGACCA,GGACCG,GGAGAA,GGAGAG,GGAGCA,GGAGCC,GGAGCG,GGAGGA,GGAGGC,GGAUGC,GGAUGG,GGGAGC,GGGAGG,GGGUGC,GGGUGG,GGUGCA,GGUGCC,GGUGGA,GGUGGC,UGAAAA,UGAAAG,UGAACA,UGAACG,UGAAGC,UGAAGG,UGACAA,UGACAG,UGACCA,UGACCG,UGAGAA,UGAGAG,UGAGCA,UGAGCG,UGAUGC,UGAUGG,UGGAGC,UGGAGG,UGGAUU,UGGUGC,UGGUGG |
| MSI1 | 1 | 2770 | 0.009950249 | 0.0040157442 | 1.309065 | AGGAGG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| DAZAP1 | 3 | 5964 | 0.019900498 | 0.0086445016 | 1.202950 | AAAAAA,AGGAAA | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUAUA,AGUAUG,AGUUAA,AGUUAG,AGUUUA,AGUUUG,GGGGGG,GUAACG,UAGGAA,UAGGAU,UAGGUA,UAGGUU,UAGUAA,UAGUAU,UAGUUA,UAGUUU,UUUUUU |
| HNRNPH3 | 4 | 7688 | 0.024875622 | 0.0111429292 | 1.158604 | AGGGAA,GGAAGG,GGAGGG,GGGAAG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| SRSF10 | 8 | 13860 | 0.044776119 | 0.0200874160 | 1.156438 | AAAAGA,AAAGAA,AAGAAG,AAGAGA,AAGGAG,AGAGAG,AGAGGG,GAGGGA | AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| HNRNPH2 | 5 | 9711 | 0.029850746 | 0.0140746688 | 1.084666 | AAGGCG,AGGGAA,GGAAGG,GGAGGG,GGGAAG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| SRSF3 | 9 | 16536 | 0.049751244 | 0.0239654858 | 1.053774 | CAUCAA,CCUCGU,CUCCAA,CUUCGA,UACAGC,UCGACA,UCUUCG,UUCGAC,UUCUUC | AACGAC,AACGAU,AACUUU,ACAUCA,ACAUCG,ACAUUC,ACCACC,ACGACG,ACGACU,ACGAUC,ACGAUU,ACUACA,ACUACG,ACUUCA,ACUUCG,ACUUCU,ACUUUA,AGAGAU,AUCAAC,AUCAUC,AUCGAC,AUCGAU,AUCGCU,AUCGUU,AUCUUC,AUUCAU,CAACGA,CACAAC,CACAUC,CACCAC,CACUAC,CACUUC,CAGAGA,CAUCAA,CAUCAC,CAUCAU,CAUCGA,CAUCGC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CGAUCG,CGCUUC,CUACAA,CUACAC,CUACAG,CUACGA,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUCAC,CUUCAG,CUUCAU,CUUCGA,CUUCUU,CUUUAU,GACUUC,GAGAUU,GAUCAA,GAUCGA,GCUUCA,GUCAAC,UACAAA,UACAAC,UACAAU,UACAGC,UACAUC,UACGAC,UACGAU,UACUUC,UAUCAA,UCAACG,UCACAA,UCAGAG,UCAUCA,UCAUCC,UCAUCG,UCAUCU,UCGACA,UCGACU,UCGAUA,UCGAUC,UCGAUU,UCGCUU,UCGUCC,UCGUUC,UCUCCA,UCUUCA,UCUUCC,UCUUCG,UGUCAA,UUACGA,UUCAAC,UUCAUC,UUCGAC,UUCGAU,UUCUCC,UUCUUC |
| ELAVL4 | 2 | 5105 | 0.014925373 | 0.0073996354 | 1.012241 | AAAAAA | AAAAAA,AUCUAA,AUUUAU,GUAUCU,UAUCUA,UAUUUA,UAUUUU,UCUAAU,UGUAUC,UUAUUU,UUGUAU,UUUAUU,UUUGUA,UUUUAU,UUUUUA,UUUUUU |
RBP on BSJ (Exon Only)
Plot
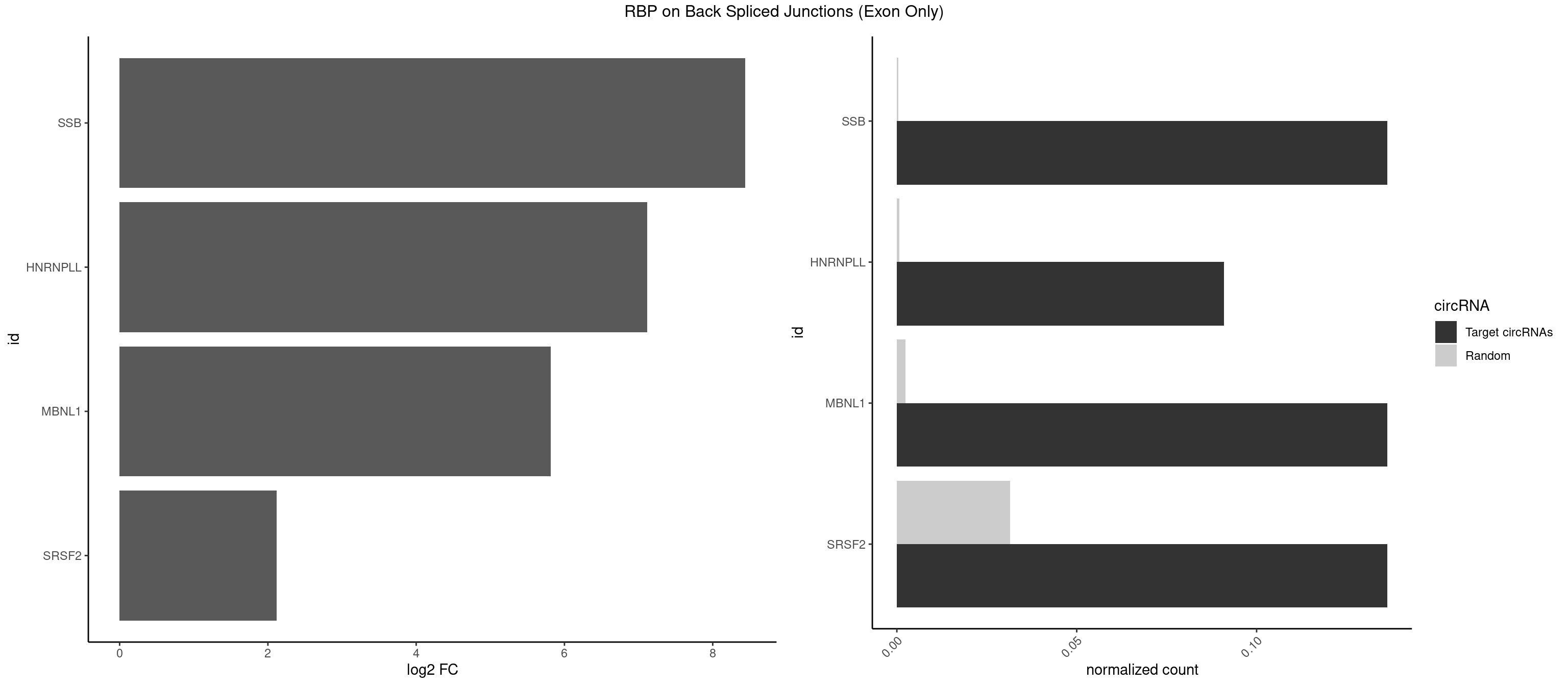
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SSB | 2 | 5 | 0.13636364 | 0.0003929788 | 8.438792 | GCUGUU,UGCUGU | CUGUUU,GCUGUU,UGCUGU |
| HNRNPLL | 1 | 9 | 0.09090909 | 0.0006549646 | 7.116864 | CACUGC | ACAAAC,ACACCA,ACAUAC,ACGACA,CACCAC,CAGACG,CAUACA,GACGAC |
| MBNL1 | 2 | 36 | 0.13636364 | 0.0024233691 | 5.814301 | CUGCUG,UGCUGU | AUGCUC,AUGCUU,CCCGCU,CCGCUA,CCGCUG,CCUGCU,CGCUGC,CGCUGU,CUGCCG,CUGCCU,CUGCGG,CUGCUC,CUGCUU,CUUGCU,GCUUGC,GCUUGU,GGCUUU,GUGCUG,UGCGGC,UGCUGC,UGCUGU,UGCUUU,UUGCCU,UUGCUC,UUGCUG,UUGCUU,UUGUGC |
| SRSF2 | 2 | 479 | 0.13636364 | 0.0314383023 | 2.116864 | GCUGUU,UGCUGU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
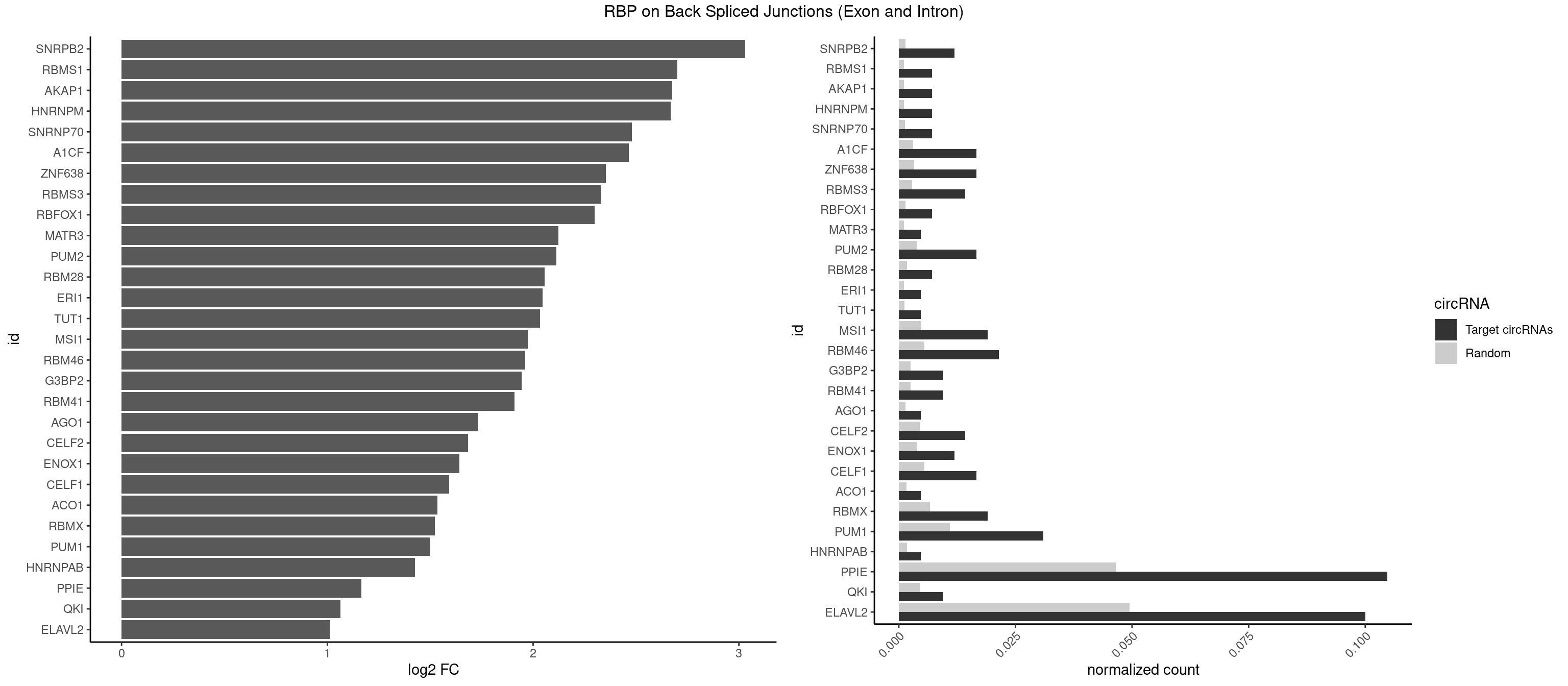
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SNRPB2 | 4 | 288 | 0.011904762 | 0.001456066 | 3.031391 | AUUGCA,UAUUGC,UGCAGU | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBMS1 | 2 | 217 | 0.007142857 | 0.001098347 | 2.701167 | AUAUAC,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| AKAP1 | 2 | 221 | 0.007142857 | 0.001118501 | 2.674935 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| HNRNPM | 2 | 222 | 0.007142857 | 0.001123539 | 2.668451 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| SNRNP70 | 2 | 253 | 0.007142857 | 0.001279726 | 2.480666 | AAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| A1CF | 6 | 598 | 0.016666667 | 0.003017936 | 2.465331 | AGUAUA,AUAAUU,UAAUUA,UGAUCA,UUAAUU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| ZNF638 | 6 | 646 | 0.016666667 | 0.003259774 | 2.354122 | GUUCUU,GUUGGU,GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBMS3 | 5 | 562 | 0.014285714 | 0.002836558 | 2.332360 | AUAUAU,CAUAUA,UAUAGC,UAUAUA,UAUAUC | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBFOX1 | 2 | 287 | 0.007142857 | 0.001451028 | 2.299426 | AGCAUG,GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| MATR3 | 1 | 216 | 0.004761905 | 0.001093309 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| PUM2 | 6 | 764 | 0.016666667 | 0.003854293 | 2.112428 | GUAUAU,UAAAUA,UACAUA,UAGAUA,UAUAUA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RBM28 | 2 | 340 | 0.007142857 | 0.001718057 | 2.055723 | AGUAGU,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| ERI1 | 1 | 228 | 0.004761905 | 0.001153769 | 2.045185 | UUUCAG | UUCAGA,UUUCAG |
| TUT1 | 1 | 230 | 0.004761905 | 0.001163845 | 2.032640 | AAAUAC | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| MSI1 | 7 | 961 | 0.019047619 | 0.004846836 | 1.974496 | AGGAAG,AGGAGG,AGUAAG,AGUUGG,UAGUAA,UAGUUA,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| RBM46 | 8 | 1091 | 0.021428571 | 0.005501814 | 1.961556 | AAUCAA,AUCAAA,AUCAUA,AUGAAA,GAUCAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| G3BP2 | 3 | 490 | 0.009523810 | 0.002473801 | 1.944809 | AGGAUG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBM41 | 3 | 502 | 0.009523810 | 0.002534260 | 1.909974 | AUACAU,UACAUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| AGO1 | 1 | 283 | 0.004761905 | 0.001430875 | 1.734641 | GUAGUA | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| CELF2 | 5 | 881 | 0.014285714 | 0.004443773 | 1.684716 | AUGUGU,GUUGUU,UGUUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| ENOX1 | 4 | 756 | 0.011904762 | 0.003813986 | 1.642167 | UAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| CELF1 | 6 | 1097 | 0.016666667 | 0.005532044 | 1.591081 | GUUUGU,UGUGUU,UGUUGU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| ACO1 | 1 | 325 | 0.004761905 | 0.001642483 | 1.535660 | CAGUGA | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| RBMX | 7 | 1316 | 0.019047619 | 0.006635429 | 1.521349 | AAGGAA,AGGAAG,AUCAAA,GAAGGA,GGAAGG,UCAAAA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| PUM1 | 12 | 2172 | 0.030952381 | 0.010948206 | 1.499356 | AAUAUU,CUUGUA,GUAAUA,GUAUAU,UAAAUA,UAAUGU,UACAUA,UAGAUA,UAUAUA,UGUAAU,UGUAUA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.001773478 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| PPIE | 43 | 9262 | 0.104761905 | 0.046669690 | 1.166556 | AAAAAU,AAAAUA,AAAUAU,AAUAAU,AAUAUU,AAUUUU,AUAAUU,AUAUAU,AUAUUA,AUAUUU,AUUAUA,AUUAUU,AUUUUA,AUUUUU,UAAAAA,UAAAUA,UAAUAA,UAAUUA,UAAUUU,UAUAUA,UAUAUU,UAUUAU,UAUUUU,UUAAAU,UUAAUU,UUAUAU,UUAUUA,UUUAAU,UUUAUA,UUUUAA | AAAAAA,AAAAAU,AAAAUA,AAAAUU,AAAUAA,AAAUAU,AAAUUA,AAAUUU,AAUAAA,AAUAAU,AAUAUA,AAUAUU,AAUUAA,AAUUAU,AAUUUA,AAUUUU,AUAAAA,AUAAAU,AUAAUA,AUAAUU,AUAUAA,AUAUAU,AUAUUA,AUAUUU,AUUAAA,AUUAAU,AUUAUA,AUUAUU,AUUUAA,AUUUAU,AUUUUA,AUUUUU,UAAAAA,UAAAAU,UAAAUA,UAAAUU,UAAUAA,UAAUAU,UAAUUA,UAAUUU,UAUAAA,UAUAAU,UAUAUA,UAUAUU,UAUUAA,UAUUAU,UAUUUA,UAUUUU,UUAAAA,UUAAAU,UUAAUA,UUAAUU,UUAUAA,UUAUAU,UUAUUA,UUAUUU,UUUAAA,UUUAAU,UUUAUA,UUUAUU,UUUUAA,UUUUAU,UUUUUA,UUUUUU |
| QKI | 3 | 904 | 0.009523810 | 0.004559653 | 1.062615 | AUCAUA,UCAUAU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| ELAVL2 | 41 | 9826 | 0.100000000 | 0.049511286 | 1.014171 | AAUUUU,AUAUUU,AUUAUU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUUCU,CUUUUA,UAAUUU,UAGUUA,UAUAUA,UAUAUU,UAUUAU,UAUUUG,UAUUUU,UCUUUU,UUAAUU,UUAGUU,UUAUAC,UUAUAU,UUAUUG,UUCUUU,UUGUAU,UUUAAU,UUUACU,UUUAUA,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUUG | AAUUUA,AAUUUG,AAUUUU,AUACUU,AUAUUU,AUUAUU,AUUUAA,AUUUAC,AUUUAG,AUUUAU,AUUUUA,AUUUUC,AUUUUG,AUUUUU,CAUUUU,CUUAUA,CUUUAA,CUUUCU,CUUUUA,CUUUUU,GAUUUA,GAUUUU,GUAUUG,GUUUUA,GUUUUC,GUUUUU,UAAGUU,UAAUUU,UACUUU,UAGUUA,UAUACU,UAUAUA,UAUAUU,UAUGUU,UAUUAU,UAUUGA,UAUUGU,UAUUUA,UAUUUG,UAUUUU,UCAUUU,UCUUAU,UCUUUU,UGAUUU,UGUAUU,UUAAGU,UUAAUU,UUACUU,UUAGUU,UUAUAC,UUAUAU,UUAUGU,UUAUUG,UUAUUU,UUCAUU,UUCUUU,UUGAUU,UUGUAU,UUUAAG,UUUAAU,UUUACU,UUUAGU,UUUAUA,UUUAUG,UUUAUU,UUUCAU,UUUCUU,UUUGAU,UUUUAA,UUUUAG,UUUUAU,UUUUUA,UUUUUC,UUUUUG,UUUUUU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.