circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000004487:+:1:23030468:23059167
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000004487:+:1:23030468:23059167 | ENSG00000004487 | MSTRG.523.27 | + | 1 | 23030469 | 23059167 | 816 | GUAGAGUACAGAGAGAUGGAUGAAAGCUUGGCCAACCUCUCAGAAGAUGAGUAUUAUUCAGAAGAAGAGAGAAAUGCCAAAGCAGAGAAGGAAAAGAAGCUUCCCCCACCACCCCCUCAAGCCCCACCUGAGGAAGAAAAUGAAAGUGAGCCUGAAGAACCAUCGGGGCAAGCAGGAGGACUUCAAGACGACAGUUCUGGAGGGUAUGGAGACGGCCAAGCAUCAGGUGUGGAGGGCGCAGCUUUCCAGAGCCGACUUCCUCAUGACCGGAUGACUUCUCAAGAAGCAGCCUGUUUUCCAGAUAUUAUCAGUGGACCACAACAGACCCAGAAGGUUUUUCUUUUCAUUAGAAACCGCACACUGCAGUUGUGGUUGGAUAAUCCAAAGAUUCAGCUGACAUUUGAGGCUACUCUCCAACAAUUAGAAGCACCUUAUAACAGUGAUACUGUGCUUGUCCACCGAGUUCACAGUUAUUUAGAGCGUCAUGGUCUUAUCAACUUCGGCAUCUAUAAGAGGAUAAAACCCCUACCAACUAAAAAGACAGGAAAGGUAAUUAUUAUAGGCUCUGGGGUCUCAGGCUUGGCAGCAGCUCGACAGUUACAAAGUUUUGGAAUGGAUGUCACACUUUUGGAAGCCAGGGAUCGUGUGGGUGGACGAGUUGCCACAUUUCGCAAAGGAAACUAUGUAGCUGAUCUUGGAGCCAUGGUGGUAACAGGUCUUGGAGGGAAUCCUAUGGCUGUGGUCAGCAAACAAGUAAAUAUGGAACUGGCCAAGAUCAAGCAAAAAUGCCCACUUUAUGAAGCCAACGGACAAGCUGUAGAGUACAGAGAGAUGGAUGAAAGCUUGGCCAACCUCUCAGAAGAUGA | circ |
| ENSG00000004487:+:1:23030468:23059167 | ENSG00000004487 | MSTRG.523.27 | + | 1 | 23030469 | 23059167 | 22 | ACGGACAAGCUGUAGAGUACAG | bsj |
| ENSG00000004487:+:1:23030468:23059167 | ENSG00000004487 | MSTRG.523.27 | + | 1 | 23030269 | 23030478 | 210 | GAACCUGAACUGACACAAGAUUGUUUCAUGAGGGCUUUGUUUUUAAUAACUACCUAUUAUGAGCUGCCUUGUUCCUUGUUGUAUGCUUACUUCCCUUAAAAUGUGAGGUUAACAAUUUACAUUGACUGAUGGUUCCAAAUAGAGUCCCUAAUUUUAAAUGUUCACUGCAUUUAGCUUUCCCUGGUAUGAUCUCCAUAUAGGUAGAGUACA | ie_up |
| ENSG00000004487:+:1:23030468:23059167 | ENSG00000004487 | MSTRG.523.27 | + | 1 | 23059158 | 23059367 | 210 | CGGACAAGCUGUAAGUCGAGGACAAACAGAACUUUCAACAUUUCUUUGGUUCACUUGAAAACAAUUUUAGGAGAAUGGUAUGGAUGAGCAUAUACAUAUAUACACAUAUAGAGGUGUAUAUAUAUAUAAACUGAGGGUAGAGAUGAUGUGAUGUUAUUCUGGUUGUUUCUUAACUCUUGAGCUAUGUAGGAAAGGAUUAUUGUAUAUAUA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
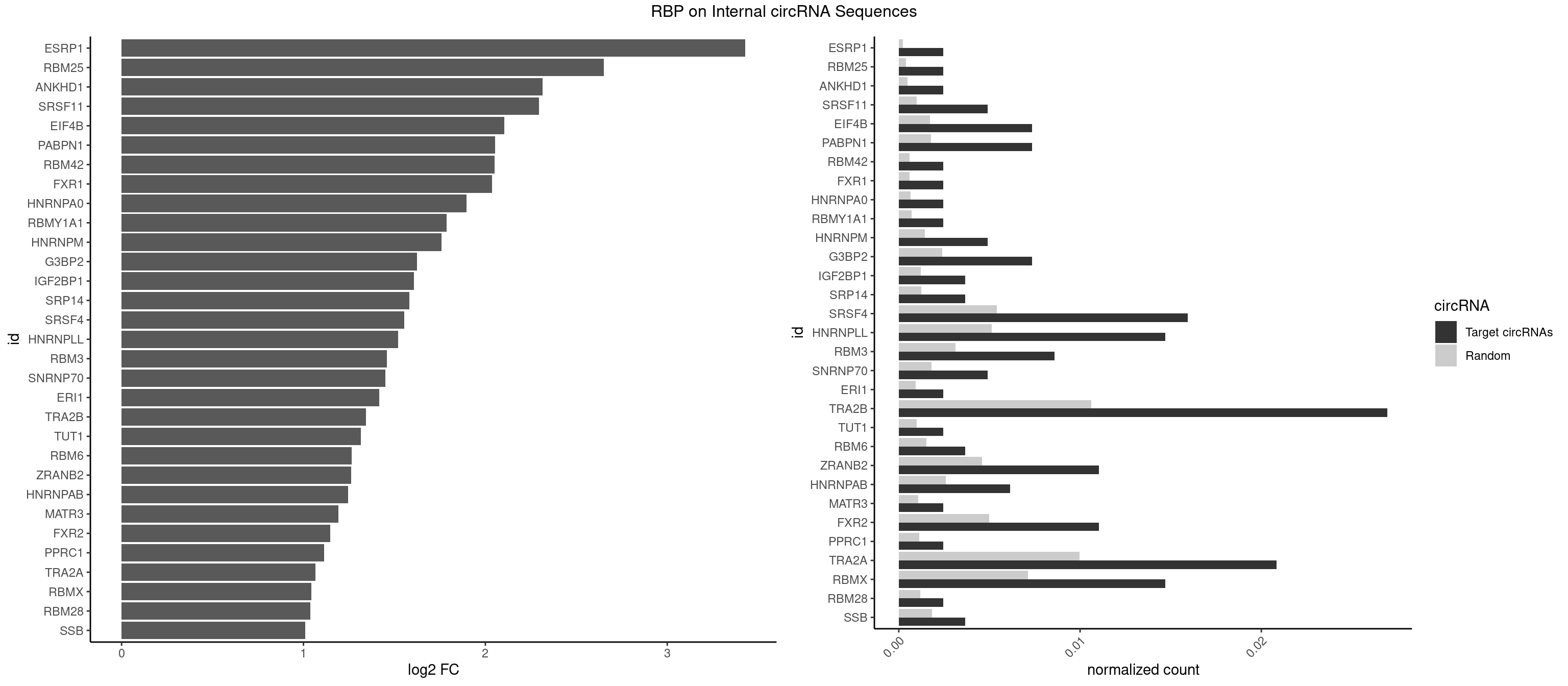
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.002450980 | 0.0002275250 | 3.429262 | AGGGAU | AGGGAU |
| RBM25 | 1 | 268 | 0.002450980 | 0.0003898359 | 2.652420 | AUCGGG | AUCGGG,CGGGCA,UCGGGC |
| ANKHD1 | 1 | 339 | 0.002450980 | 0.0004927293 | 2.314492 | AGACGA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| SRSF11 | 3 | 688 | 0.004901961 | 0.0009985015 | 2.295522 | AAGAAG | AAGAAG |
| EIF4B | 5 | 1179 | 0.007352941 | 0.0017100607 | 2.104274 | CUUGGA,GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| PABPN1 | 5 | 1222 | 0.007352941 | 0.0017723764 | 2.052636 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBM42 | 1 | 407 | 0.002450980 | 0.0005912752 | 2.051457 | AACUAA | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 1 | 411 | 0.002450980 | 0.0005970720 | 2.037382 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| HNRNPA0 | 1 | 453 | 0.002450980 | 0.0006579386 | 1.897334 | AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| RBMY1A1 | 1 | 489 | 0.002450980 | 0.0007101099 | 1.787245 | CAAGAC | ACAAGA,CAAGAC |
| HNRNPM | 3 | 999 | 0.004901961 | 0.0014492040 | 1.758098 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| G3BP2 | 5 | 1644 | 0.007352941 | 0.0023839405 | 1.624973 | AGGAUA,GGAUAA,GGAUGA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| IGF2BP1 | 2 | 831 | 0.003676471 | 0.0012057377 | 1.608405 | AAGCAC,AGCACC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SRP14 | 2 | 847 | 0.003676471 | 0.0012289250 | 1.580925 | CUGUAG,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| SRSF4 | 12 | 3740 | 0.015931373 | 0.0054214720 | 1.555114 | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| HNRNPLL | 11 | 3534 | 0.014705882 | 0.0051229360 | 1.521351 | ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACCAC,CACUGC,GACGAC,GCAAAC,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| RBM3 | 6 | 2152 | 0.008578431 | 0.0031201361 | 1.459105 | AAACUA,AAGACG,AGACGA,GAAACU,GAGACG,GAUACU | AAAACG,AAAACU,AAACGA,AAACUA,AAGACG,AAGACU,AAUACG,AAUACU,AGACGA,AGACUA,AUACGA,AUACUA,GAAACG,GAAACU,GAGACG,GAGACU,GAUACG,GAUACU |
| SNRNP70 | 3 | 1237 | 0.004901961 | 0.0017941145 | 1.450087 | AUCAAG,GAUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| ERI1 | 1 | 632 | 0.002450980 | 0.0009173461 | 1.417821 | UUCAGA | UUCAGA,UUUCAG |
| TRA2B | 21 | 7329 | 0.026960784 | 0.0106226650 | 1.343717 | AAAGAA,AAGAAC,AAGAAG,AAGGAA,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAGAA,GAAGGA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| TUT1 | 1 | 678 | 0.002450980 | 0.0009840095 | 1.316615 | GAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| RBM6 | 2 | 1054 | 0.003676471 | 0.0015289102 | 1.265818 | AAUCCA,AUCCAA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ZRANB2 | 8 | 3173 | 0.011029412 | 0.0045997733 | 1.261721 | AAAGGU,AGGGAA,AGGUAA,AGGUCU,AGGUUU,GGUGGU,UGGUAA,UGGUGG | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| HNRNPAB | 4 | 1782 | 0.006127451 | 0.0025839306 | 1.245720 | AAAGAC,AAGACA,ACAAAG,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| MATR3 | 1 | 739 | 0.002450980 | 0.0010724109 | 1.192501 | AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| FXR2 | 8 | 3434 | 0.011029412 | 0.0049780156 | 1.147713 | AGACAG,AGACGA,AGACGG,GACAAG,GACAGG,GACGAG,GGACAA,GGACGA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| PPRC1 | 1 | 780 | 0.002450980 | 0.0011318283 | 1.114704 | GGGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| TRA2A | 16 | 6871 | 0.020833333 | 0.0099589296 | 1.064831 | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAGA,AGAGGA,AGGAAG,GAAGAA,GAAGAG,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| RBMX | 11 | 4925 | 0.014705882 | 0.0071387787 | 1.042644 | AAGAAG,AAGGAA,AAGUAA,AGAAGG,AGGAAG,GAAGGA,GUAACA | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| RBM28 | 1 | 822 | 0.002450980 | 0.0011926949 | 1.039134 | UGUAGA | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| SSB | 2 | 1260 | 0.003676471 | 0.0018274462 | 1.008493 | CUGUUU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
RBP on BSJ (Exon Only)
Plot
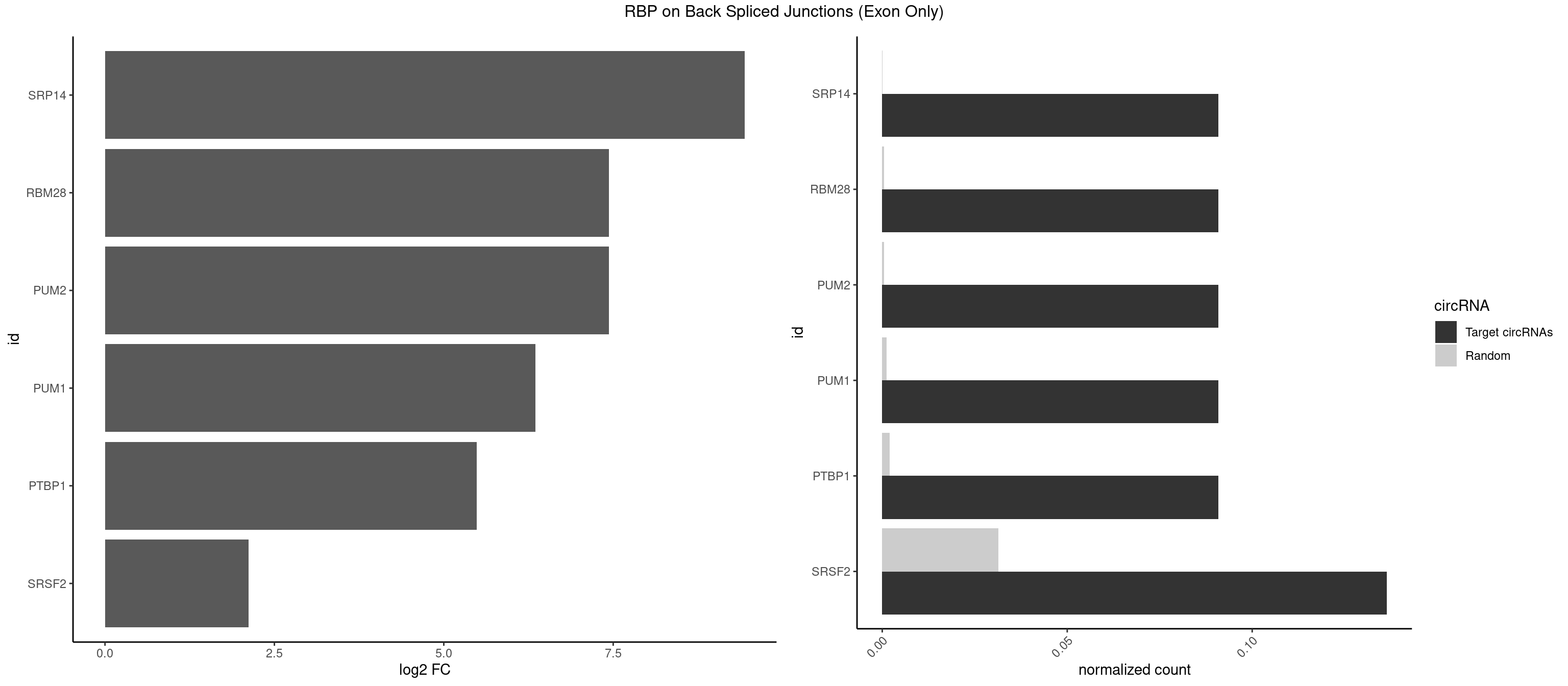
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRP14 | 1 | 1 | 0.09090909 | 0.0001309929 | 9.438792 | CUGUAG | GCCUGU |
| PUM2 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | UGUAGA | GUAAAU,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA |
| RBM28 | 1 | 7 | 0.09090909 | 0.0005239717 | 7.438792 | UGUAGA | AGUAGG,AGUAGU,UGUAGA,UGUAGG,UGUAGU |
| PUM1 | 1 | 16 | 0.09090909 | 0.0011134399 | 6.351329 | UGUAGA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAUAA,CAGAAU,GUAAAU,GUCCAG,UAAAUA,UAUAUA,UGUAAA,UGUAGA,UGUAUA,UUAAUG |
| PTBP1 | 1 | 30 | 0.09090909 | 0.0020303904 | 5.484596 | AGCUGU | AGCUGU,CAUCUU,CCAUCU,CCUCUU,CUAUCU,CUCUUA,CUCUUC,CUCUUG,CUCUUU,CUUUCU,GGCUCC,GUCUUU,UACUUU,UAUUUU,UCCUCU,UCUAUC,UCUCUU,UCUUCU,UCUUUC,UUACUU,UUCUUC,UUCUUG,UUUCUU |
| SRSF2 | 2 | 479 | 0.13636364 | 0.0314383023 | 2.116864 | AAGCUG,AGCUGU | AAAAGA,AAAGAG,AAGAGA,AAGCAG,AAGCUG,AAGGCG,AAUACC,AAUGCU,ACCACC,ACCACU,ACCAGC,ACCAGU,ACCCCC,ACCCCU,ACUCAA,AGAAGA,AGAAGC,AGAAUA,AGAAUG,AGAGAA,AGAGAU,AGAGGA,AGAGGU,AGAGUA,AGAGUG,AGAGUU,AGAUAA,AGAUCC,AGAUGC,AGCACU,AGCAGA,AGCAGU,AGCCGA,AGCCUC,AGCGGA,AGCUGU,AGGAAG,AGGAGA,AGGAGC,AGGAGG,AGGAGU,AGGCAG,AGGCGU,AGGCUG,AGGGUA,AGUAGG,AGUAGU,AGUGAC,AGUGUU,AUGAUG,AUGCUG,AUGGAG,AUUAGU,AUUCCU,AUUGAU,CAGAGA,CAGAGG,CAGAGU,CAGUAG,CAGUGG,CCACCA,CCAGAU,CCAGCC,CCAGCU,CCAGGG,CCAGGU,CCAGUC,CCAGUG,CCAGUU,CCCGCU,CCCGUG,CCGCUA,CCGGUG,CCUCCG,CCUGCG,CCUGCU,CCUGUU,CGAACG,CGAGGA,CGAGUA,CGAGUG,CGAGUU,CGCAGU,CGCUGC,CGUAAG,CGUGCG,CUACCG,CUAGAA,CUCAAG,CUCCAA,CUCCUG,CUCGUG,CUGAUG,GAAAGG,GAAGAA,GAAGCG,GAAGGC,GAAUAC,GAAUCC,GACCCC,GACGGA,GACUCA,GACUGU,GAGAAG,GAGAAU,GAGAGA,GAGAGU,GAGAUA,GAGAUG,GAGCAC,GAGCAG,GAGCUG,GAGGAA,GAGGAC,GAGGAG,GAGUGA,GAUCCC,GAUCCG,GAUGAU,GAUGCU,GAUGGA,GAUUAG,GAUUGA,GCACUG,GCAGAG,GCAGGG,GCAGGU,GCAGUA,GCAGUG,GCCACC,GCCACU,GCCCAC,GCCGAG,GCCGCC,GCCGUU,GCCUCA,GCCUCC,GCGUGU,GCUGUU,GGAACC,GGAAGG,GGAAUG,GGACCG,GGACGC,GGAGAA,GGAGAG,GGAGAU,GGAGCG,GGAGGA,GGAGUG,GGAGUU,GGAUCC,GGAUGG,GGCAGU,GGCCAC,GGCCGC,GGCCUC,GGCGUG,GGCUCC,GGCUCG,GGCUGA,GGCUGC,GGGAAU,GGGAGC,GGGCAG,GGGUAA,GGGUAC,GGGUAG,GGGUAU,GGGUGA,GGUACG,GGUAGG,GGUCAG,GGUUAC,GGUUCC,GGUUGG,GUAAGC,GUAGGC,GUCAGU,GUCGCC,GUCUAA,GUGCAG,GUUAAU,GUUCCC,GUUCCU,GUUCGA,GUUCUG,GUUGGC,GUUUCG,UAAGCU,UACGAG,UACGUG,UAGGCU,UAUGCU,UCACCG,UCAGUG,UCCAGA,UCCAGC,UCCAGG,UCCAGU,UCCUGC,UCCUGU,UCGAGU,UCGUGC,UGAGCU,UGAUCG,UGAUGG,UGCAGA,UGCAGU,UGCCGU,UGCGGU,UGCUGU,UGGAGA,UGGAGG,UGGAGU,UGGCAG,UGUUCC,UUAAUG,UUACUG,UUAGUG,UUCCAG,UUCCCG,UUCCUA,UUCCUG,UUCGAG,UUGUUG |
RBP on BSJ (Exon and Intron)
Plot
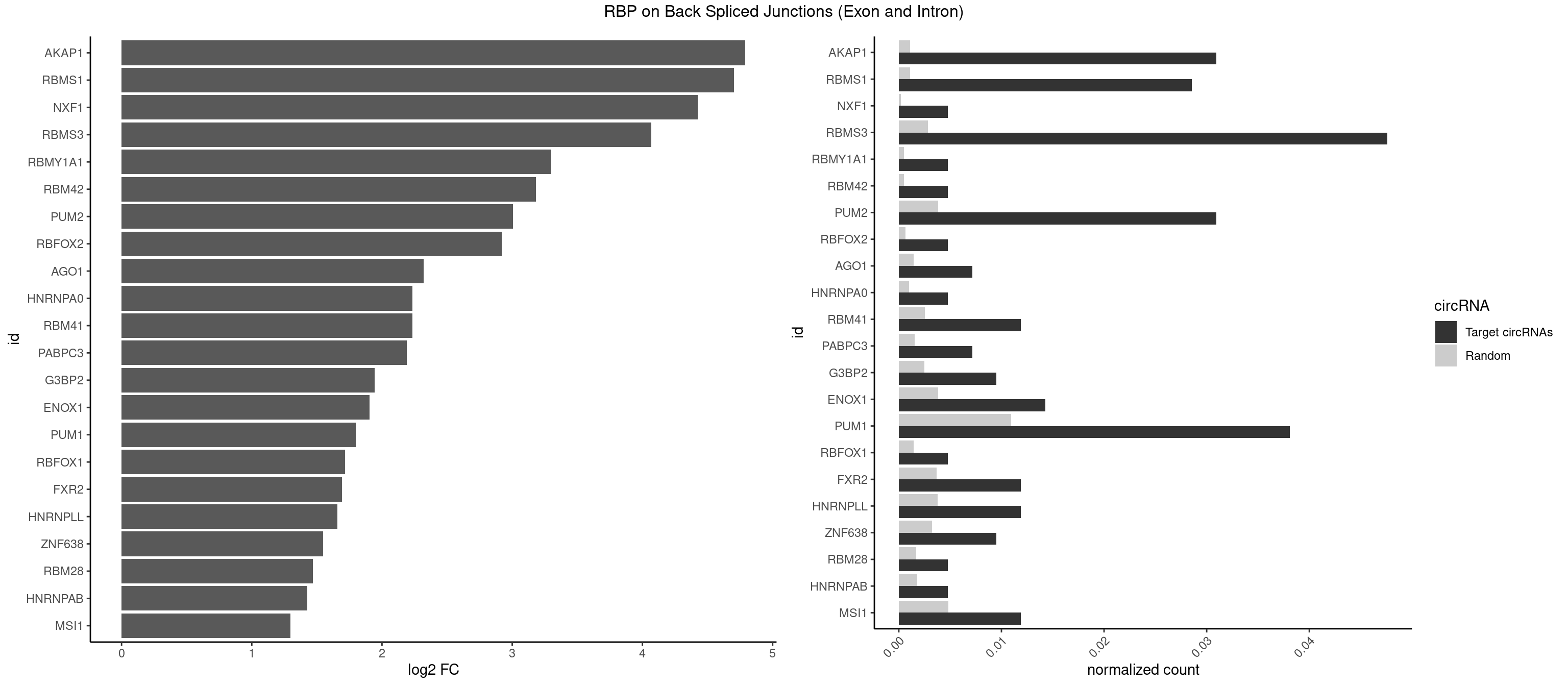
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| AKAP1 | 12 | 221 | 0.030952381 | 0.0011185006 | 4.790412 | AUAUAU,UAUAUA | AUAUAU,UAUAUA |
| RBMS1 | 11 | 217 | 0.028571429 | 0.0010983474 | 4.701167 | AUAUAC,AUAUAG,UAUAUA | AUAUAC,AUAUAG,GAUAUA,UAUAUA |
| NXF1 | 1 | 43 | 0.004761905 | 0.0002216848 | 4.424957 | AACCUG | AACCUG |
| RBMS3 | 19 | 562 | 0.047619048 | 0.0028365578 | 4.069326 | AUAUAG,AUAUAU,CAUAUA,UAUAGA,UAUAUA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| RBMY1A1 | 1 | 95 | 0.004761905 | 0.0004836759 | 3.299426 | ACAAGA | ACAAGA,CAAGAC |
| RBM42 | 1 | 103 | 0.004761905 | 0.0005239823 | 3.183949 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| PUM2 | 12 | 764 | 0.030952381 | 0.0038542926 | 3.005512 | GUAUAU,UACAUA,UAUAUA,UGUAUA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGACUG | UGACUG,UGCAUG |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | AGGUAG,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| RBM41 | 4 | 502 | 0.011904762 | 0.0025342604 | 2.231902 | AUACAU,UACAUU,UUACAU,UUACUU | AUACAU,AUACUU,UACAUG,UACAUU,UACUUG,UACUUU,UUACAU,UUACUU |
| PABPC3 | 2 | 310 | 0.007142857 | 0.0015669085 | 2.188580 | AAAACA,GAAAAC | AAAAAC,AAAACA,AAAACC,GAAAAC |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUU,GGAUGA,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| ENOX1 | 5 | 756 | 0.014285714 | 0.0038139863 | 1.905202 | AGGACA,AGUACA,CGGACA,UAUACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| PUM1 | 15 | 2172 | 0.038095238 | 0.0109482064 | 1.798916 | AAUGUU,AUUGUA,GUAUAU,UACAUA,UAUAUA,UGUAUA,UUUAAU | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | UGACUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| FXR2 | 4 | 730 | 0.011904762 | 0.0036829907 | 1.692589 | GACAAA,GACAAG,GGACAA | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPLL | 4 | 748 | 0.011904762 | 0.0037736800 | 1.657495 | ACAAAC,ACUGCA,CAAACA,CACUGC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUGU,GUUGUU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RBM28 | 1 | 340 | 0.004761905 | 0.0017180572 | 1.470761 | UGUAGG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGUAG,UAGGAA,UAGGAG,UAGGUA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.