circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000214331:-:16:74353776:74360891
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000214331:-:16:74353776:74360891 | ENSG00000214331 | MSTRG.13017.3 | - | 16 | 74353777 | 74360891 | 393 | UUUGAGUUCCUGAGAUCUAGUUGGUGAGAGACAUGAUGUUCUACUGGUUGCUGUCGAUUGUUGGAAGACAAAGAGCCAGCCCAGGAUGGCAGAACUGGUCCUCUGCAAGAAACAGCGCAUCAGCUGCCGAGGCGCGUUCCAUGGCCCUGCCCACCCAGGCACAGGUGGUCGUCUGUGGAGGUGGAAUCACGGGCACUUCUGUGGCCCAUCACCAAUCCAAAAUGGGGUGGAAGGAUAUUGUCCUUUUGGAGCAGGGCAGGCUGGCUGCUGGCUCUACCAGGUUCUGUGCUGGCAUCCUGAGCACUGCCAGGCACUUGACCAUUGAGCAGAAGAUGGCAGACUACUCAAACAAACUCUACCAUCAGUUAGAGCAAGAAACAGGGAUCCGAACAGUUUGAGUUCCUGAGAUCUAGUUGGUGAGAGACAUGAUGUUCUACUGGUUG | circ |
| ENSG00000214331:-:16:74353776:74360891 | ENSG00000214331 | MSTRG.13017.3 | - | 16 | 74353777 | 74360891 | 22 | GAUCCGAACAGUUUGAGUUCCU | bsj |
| ENSG00000214331:-:16:74353776:74360891 | ENSG00000214331 | MSTRG.13017.3 | - | 16 | 74360882 | 74361091 | 210 | AGGAUGGUCUCGAUCUCCUGACCUCGUGAUCCGCCCGCCUCGGCCUCCCAGAGUGCUGGGAUUACAGGUGUGAGGCACCGCGCCCGGCCACCCUCAAAUAUUUGCUGAAUGAAUGGCUAUCGUUCUUUGUCAAAUCCCUUUCUCAUUAAUCCCAUGCACUGAGUUGAAUGGCAUGAAAUAUGUUUGGUUUCUCUGUCUAGUUUGAGUUCC | ie_up |
| ENSG00000214331:-:16:74353776:74360891 | ENSG00000214331 | MSTRG.13017.3 | - | 16 | 74353577 | 74353786 | 210 | AUCCGAACAGGUAAGCAAGUGUCUUCCAUUUAUUGCUUUGCCUGUCCCUGAGAUUGUUAUAUUCAUUUCUGUAUUUGGUCCUCUGCCAGGUGUGCCUACUGCUUUGGUAAUUAAAACUUGGGUUGGCCGGUUUUAAACAUGGUGGUUCAUGCCUGUAAUCCCAGCACUUUGGGAGGCCGAGGCAGGCGGAUCACGAGGUCAGGAGAUCGA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
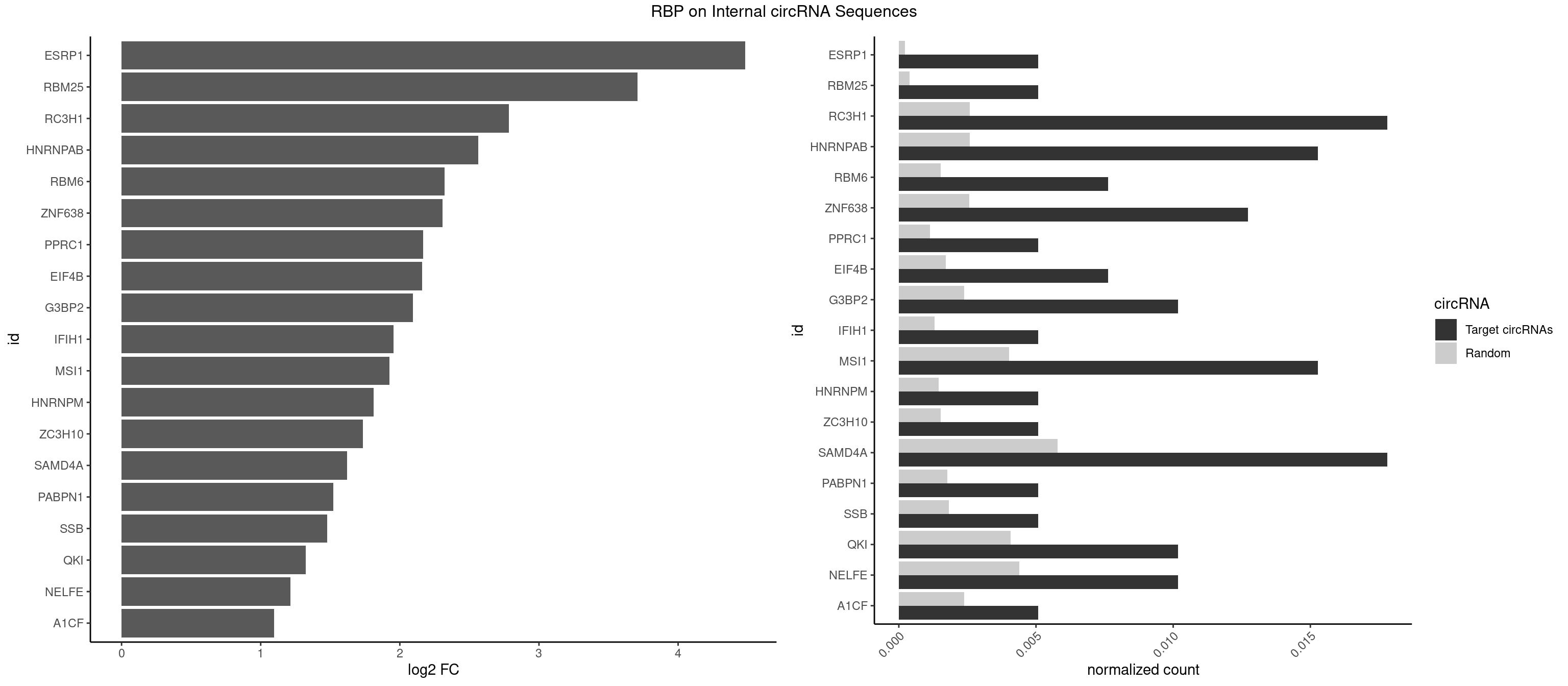
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| ESRP1 | 1 | 156 | 0.005089059 | 0.0002275250 | 4.483302 | AGGGAU | AGGGAU |
| RBM25 | 1 | 268 | 0.005089059 | 0.0003898359 | 3.706460 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| RC3H1 | 6 | 1786 | 0.017811705 | 0.0025897275 | 2.781953 | CUUCUG,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| HNRNPAB | 5 | 1782 | 0.015267176 | 0.0025839306 | 2.562794 | AAGACA,ACAAAG,AGACAA,CAAAGA,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| RBM6 | 2 | 1054 | 0.007633588 | 0.0015289102 | 2.319858 | AAUCCA,AUCCAA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| ZNF638 | 4 | 1773 | 0.012722646 | 0.0025708878 | 2.307060 | GGUUCU,GUUGGU,UGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| PPRC1 | 1 | 780 | 0.005089059 | 0.0011318283 | 2.168744 | GGCGCG | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| EIF4B | 2 | 1179 | 0.007633588 | 0.0017100607 | 2.158314 | GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| G3BP2 | 3 | 1644 | 0.010178117 | 0.0023839405 | 2.094051 | AGGAUA,AGGAUG,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| IFIH1 | 1 | 904 | 0.005089059 | 0.0013115296 | 1.956148 | GGCCCU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| MSI1 | 5 | 2770 | 0.015267176 | 0.0040157442 | 1.926694 | AGGUGG,AGUUAG,AGUUGG,UAGUUG | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| HNRNPM | 1 | 999 | 0.005089059 | 0.0014492040 | 1.812138 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| ZC3H10 | 1 | 1053 | 0.005089059 | 0.0015274610 | 1.736263 | CAGCGC | CAGCGA,CAGCGC,CCAGCG,CGAGCG,GAGCGA,GAGCGC,GCAGCG,GGAGCG |
| SAMD4A | 6 | 3992 | 0.017811705 | 0.0057866714 | 1.622020 | CGGGCA,CUGGCA,CUGGUC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPN1 | 1 | 1222 | 0.005089059 | 0.0017723764 | 1.521714 | AGAAGA | AAAAGA,AGAAGA |
| SSB | 1 | 1260 | 0.005089059 | 0.0018274462 | 1.477570 | UGCUGU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| QKI | 3 | 2805 | 0.010178117 | 0.0040664663 | 1.323623 | CUACUC,UACUCA,UCUACU | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| NELFE | 3 | 3028 | 0.010178117 | 0.0043896388 | 1.213297 | CUGGCU,CUGGUU | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| A1CF | 1 | 1642 | 0.005089059 | 0.0023810421 | 1.095806 | AUCAGU | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
RBP on BSJ (Exon Only)
Plot
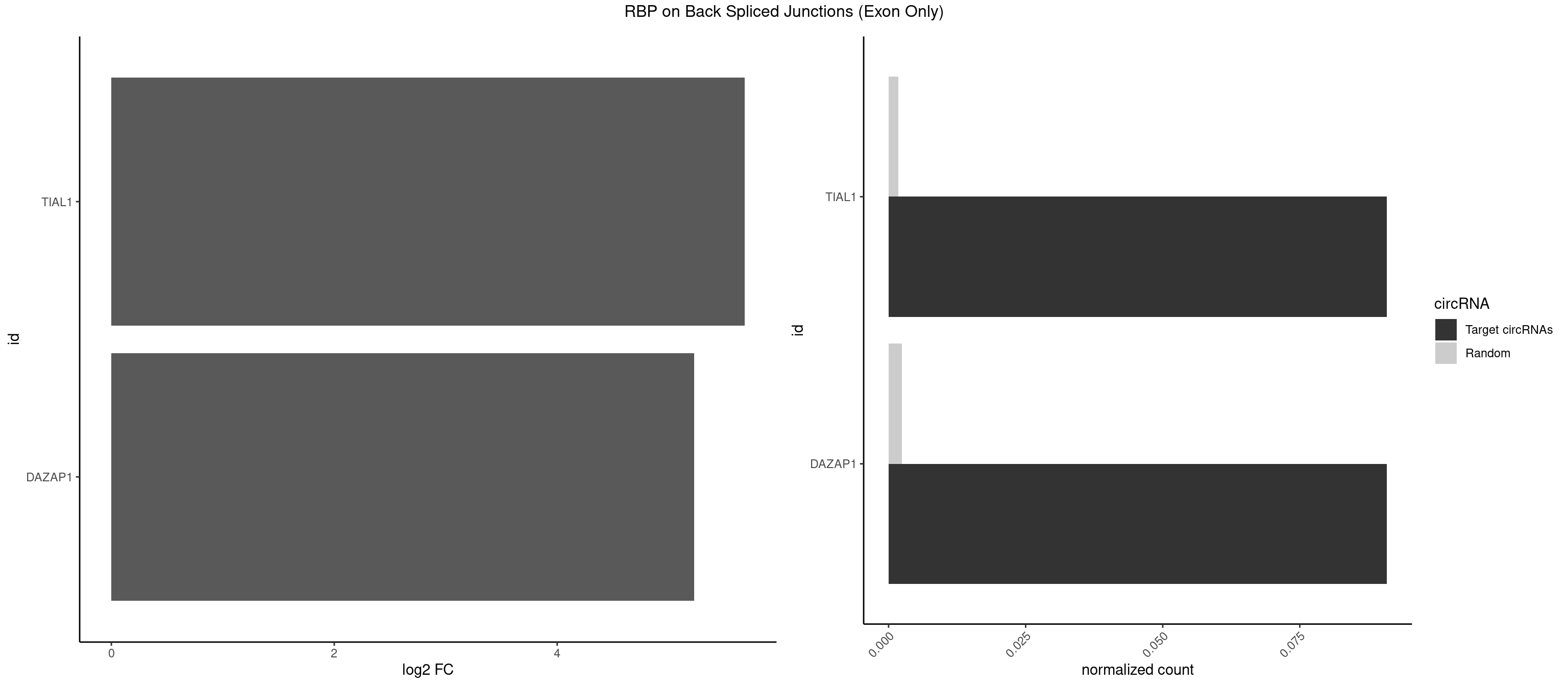
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| TIAL1 | 1 | 26 | 0.09090909 | 0.001768405 | 5.683904 | CAGUUU | AAAUUU,AAUUUU,AGUUUU,AUUUUG,CAGUUU,GUUUUU,UAAAUU,UAUUUU,UCAGUU,UUAAAU,UUCAGU,UUUCAG |
| DAZAP1 | 1 | 36 | 0.09090909 | 0.002423369 | 5.229338 | AGUUUG | AAAAAA,AAUUUA,AGAUAU,AGGAAA,AGGAAG,AGGAUA,AGGAUG,AGGUAA,AGGUAG,AGGUUA,AGGUUG,AGUAAA,AGUAAG,AGUAGG,AGUUAA,AGUUUG,GUAACG,UAGGAA,UAGGUU,UAGUUA |
RBP on BSJ (Exon and Intron)
Plot
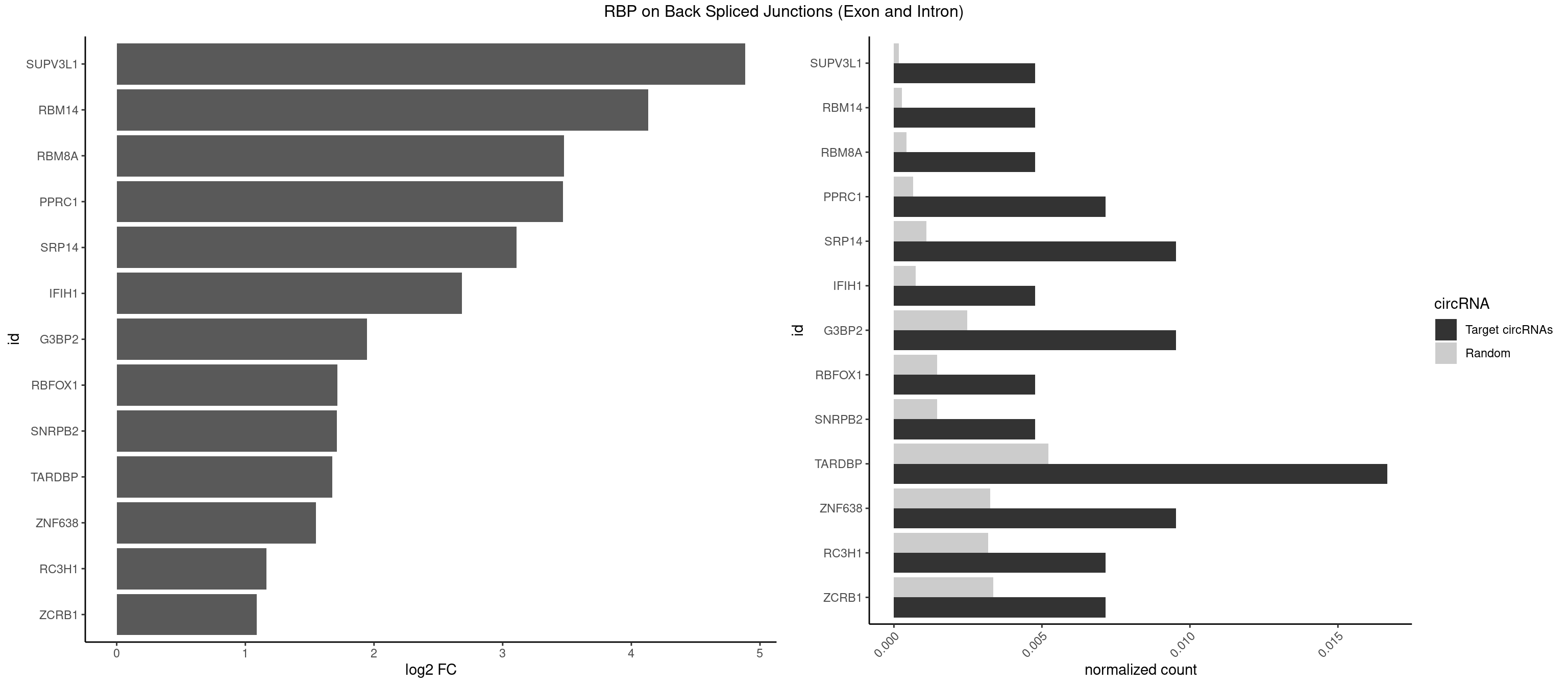
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 1 | 31 | 0.004761905 | 0.0001612253 | 4.884389 | CCGCCC | CCGCCC |
| RBM14 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | CGCGCC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
| RBM8A | 1 | 84 | 0.004761905 | 0.0004282547 | 3.474998 | CGCGCC | ACGCGC,AUGCGC,CGCGCC,CGCGCG,CGCGCU,GCGCGC,GUGCGC,UGCGCC,UGCGCG,UGCGCU |
| PPRC1 | 2 | 127 | 0.007142857 | 0.0006449012 | 3.469351 | CCGCGC,CGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| SRP14 | 3 | 218 | 0.009523810 | 0.0011033857 | 3.109602 | CCUGUA,GCCUGU | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| IFIH1 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | GCGGAU | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUG,GGAUGG,GGAUUA | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| RBFOX1 | 1 | 287 | 0.004761905 | 0.0014510278 | 1.714464 | GCAUGA | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| TARDBP | 6 | 1034 | 0.016666667 | 0.0052146312 | 1.676328 | GAAUGA,GAAUGG,UGAAUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | CGUUCU,GGUUGG,GUUCUU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| RC3H1 | 2 | 631 | 0.007142857 | 0.0031841999 | 1.165570 | UCCCUU,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | AAUUAA,GGAUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.