circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000121073_ENSG00000121104:-:17:49716164:49717714
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000121073_ENSG00000121104:-:17:49716164:49717714 | ENSG00000121073_ENSG00000121104 | MSTRG.14643.15 | - | 17 | 49716165 | 49717714 | 353 | AUCCUUGAUAUCCCUGAUGGGCACCGGGCCCCAGCUCCUCCCCAGAGUGGCAGCUGUGAUCAUCCCCUCCUCCUCCUGGAGCCUGGCAACCUUGCCAGCUCUCCUUCCAUGUCCUUGGCAUCUCCCCAGCCUUGUGGCCUGGCCAGUCAUGAGGAACAUCGGGGUGCCGCCGAGGAGCUGGCAUCCACCCCCAACGACAAAGCCUCCUCUCCAGGACACCCAGCCUUUCUUGAAGAUGGCAGCCCAUCUCCAGUCCUUGCCUUUGCUGCCUCCCCUCGACCUAAUCAUAGCUACAUCUUCAAACGGGAGCCCCCAGAAGGCUGUGAGAAAGUGCGUGUGUUUGAAGAAGCCACAUCCUUGAUAUCCCUGAUGGGCACCGGGCCCCAGCUCCUCCCCAGAGUGG | circ |
| ENSG00000121073_ENSG00000121104:-:17:49716164:49717714 | ENSG00000121073_ENSG00000121104 | MSTRG.14643.15 | - | 17 | 49716165 | 49717714 | 22 | GAAGAAGCCACAUCCUUGAUAU | bsj |
| ENSG00000121073_ENSG00000121104:-:17:49716164:49717714 | ENSG00000121073_ENSG00000121104 | MSTRG.14643.15 | - | 17 | 49717705 | 49717914 | 210 | GUGCUAUGGAAGAAACAGGAGGGAGUUUCUUCUAGAUUGAAGGGUGGCCAGGGAAAUAAAUCGCUGGAGGAAAUGGUGUUCUGUGGUUGUGUGUCAUGGAUAGAGAAUGACAGCAGCAAGGGAAAUACCCUUGGGGUCACUGUGCUGCUGCAGGCCUGAAAUACUCAGGGCUGACAGUCUUUACCGUCCUUCCCCGUUAGAUCCUUGAUA | ie_up |
| ENSG00000121073_ENSG00000121104:-:17:49716164:49717714 | ENSG00000121073_ENSG00000121104 | MSTRG.14643.15 | - | 17 | 49715965 | 49716174 | 210 | AAGAAGCCACGUGAGUACACCAAGUGGACAGGGUGUGGGAGGGGAGGGUGGGCCUACAUUCUCCCACCUUCUUCAGUCUUGGACUCUCUCUUGUCAUAGAGGUGUUGUCAACUUUAGAUGAUAAGUAUUGCCAGCUUUCCAUGAUCAAACGUGUUGACAUUACCUACUCUGAGCACUUAAAUCUUUGCUUAUCAUUAUCUCCAUAGAAAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
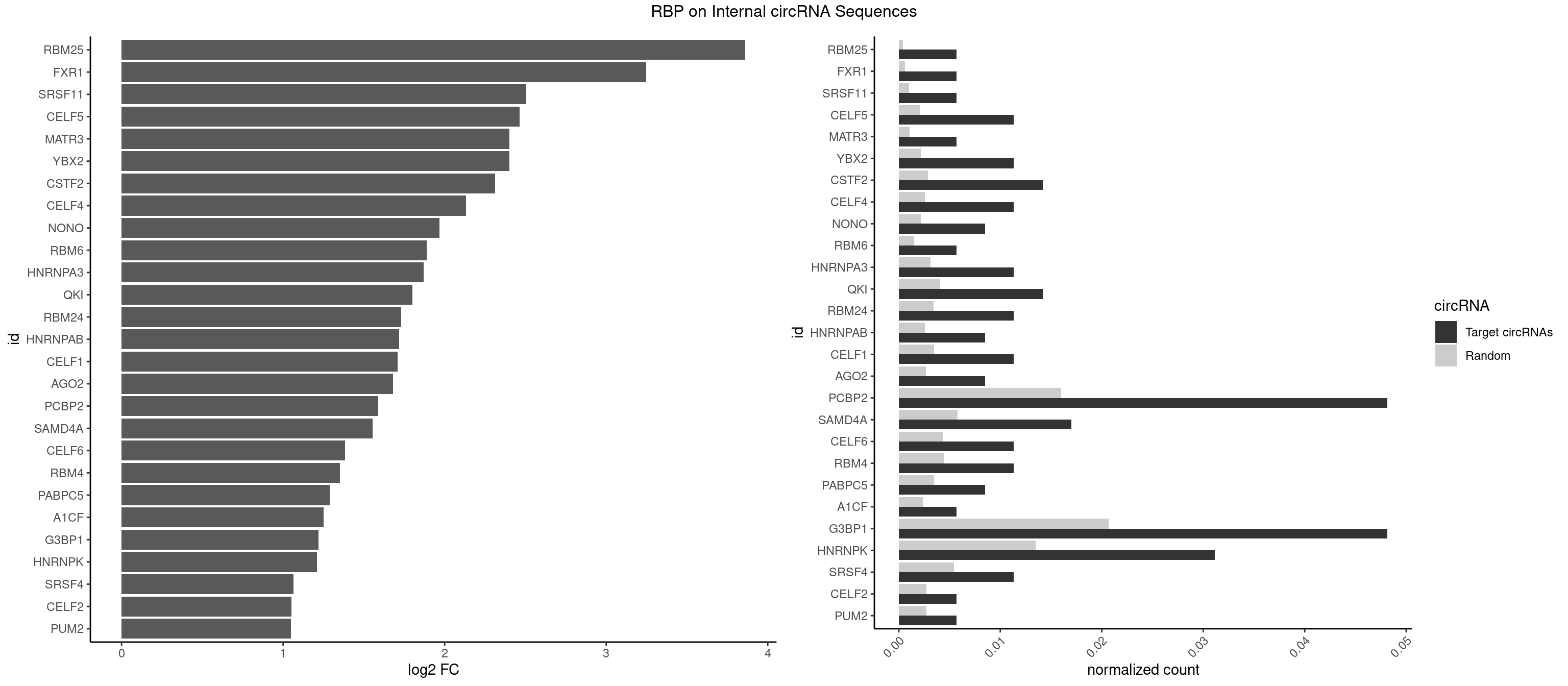
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 268 | 0.005665722 | 0.0003898359 | 3.861321 | AUCGGG | AUCGGG,CGGGCA,UCGGGC |
| FXR1 | 1 | 411 | 0.005665722 | 0.0005970720 | 3.246283 | ACGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRSF11 | 1 | 688 | 0.005665722 | 0.0009985015 | 2.504423 | AAGAAG | AAGAAG |
| CELF5 | 3 | 1415 | 0.011331445 | 0.0020520728 | 2.465178 | GUGUGU,GUGUUU,UGUGUU | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| MATR3 | 1 | 739 | 0.005665722 | 0.0010724109 | 2.401402 | CAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| YBX2 | 3 | 1480 | 0.011331445 | 0.0021462711 | 2.400428 | AACAUC,ACAUCG,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| CSTF2 | 4 | 1967 | 0.014164306 | 0.0028520334 | 2.312197 | GUGUGU,GUGUUU,UGUGUU,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF4 | 3 | 1782 | 0.011331445 | 0.0025839306 | 2.132693 | GUGUGU,GUGUUU,UGUGUU | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| NONO | 2 | 1498 | 0.008498584 | 0.0021723567 | 1.967961 | AGGAAC,GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM6 | 1 | 1054 | 0.005665722 | 0.0015289102 | 1.889756 | CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| HNRNPA3 | 3 | 2140 | 0.011331445 | 0.0031027457 | 1.868714 | AGGAGC,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| QKI | 4 | 2805 | 0.014164306 | 0.0040664663 | 1.800412 | AAUCAU,AUCAUA,CUAAUC,UAAUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| RBM24 | 3 | 2357 | 0.011331445 | 0.0034172229 | 1.729436 | AGAGUG,GAGUGG,GUGUGU | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| HNRNPAB | 2 | 1782 | 0.008498584 | 0.0025839306 | 1.717655 | ACAAAG,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| CELF1 | 3 | 2391 | 0.011331445 | 0.0034664959 | 1.708782 | GUGUGU,UGUGUU,UGUUUG | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| AGO2 | 2 | 1830 | 0.008498584 | 0.0026534924 | 1.679330 | AAAGUG,AAGUGC | AAAAAA,AAAGUG,AAGUGC,AGUGCU,GUGCUU,UAAAGU |
| PCBP2 | 16 | 11045 | 0.048158640 | 0.0160079069 | 1.589010 | CCCCCA,CCCUCC,CCUCCC,CCUCCU,CCUUCC,CUCCCC,CUUCCA,UCCCCA | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| SAMD4A | 5 | 3992 | 0.016997167 | 0.0057866714 | 1.554489 | CGGGCC,CUGGCA,CUGGCC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| CELF6 | 3 | 2997 | 0.011331445 | 0.0043447134 | 1.382999 | UGUGAG,UGUGAU,UGUGUU | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| RBM4 | 3 | 3065 | 0.011331445 | 0.0044432593 | 1.350642 | CCUUCC,UCCUUC,UUCUUG | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| PABPC5 | 2 | 2400 | 0.008498584 | 0.0034795387 | 1.288326 | AGAAAG,GAAAGU | AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| A1CF | 1 | 1642 | 0.005665722 | 0.0023810421 | 1.250667 | UGAUCA | AGUAUA,AUAAUU,AUCAGU,CAGUAU,GAUCAG,UAAUUA,UAAUUG,UCAGUA,UGAUCA,UUAAUU |
| G3BP1 | 16 | 14282 | 0.048158640 | 0.0206989801 | 1.218235 | ACCCCC,CACCGG,CAUCGG,CCACCC,CCCAUC,CCCCAG,CCCCCA,CCCCUC,CCCUCC,CCCUCG,CCGCCG | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| HNRNPK | 10 | 9304 | 0.031161473 | 0.0134848428 | 1.208425 | ACCCCC,CAUCCC,CCCCAA,CCCCCA,GCCCCC,UCCCCA,UCCCCU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| SRSF4 | 3 | 3740 | 0.011331445 | 0.0054214720 | 1.063575 | AAGAAG,GAAGAA,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| CELF2 | 1 | 1886 | 0.005665722 | 0.0027346479 | 1.050905 | GUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| PUM2 | 1 | 1890 | 0.005665722 | 0.0027404447 | 1.047850 | UACAUC | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
RBP on BSJ (Exon Only)
Plot
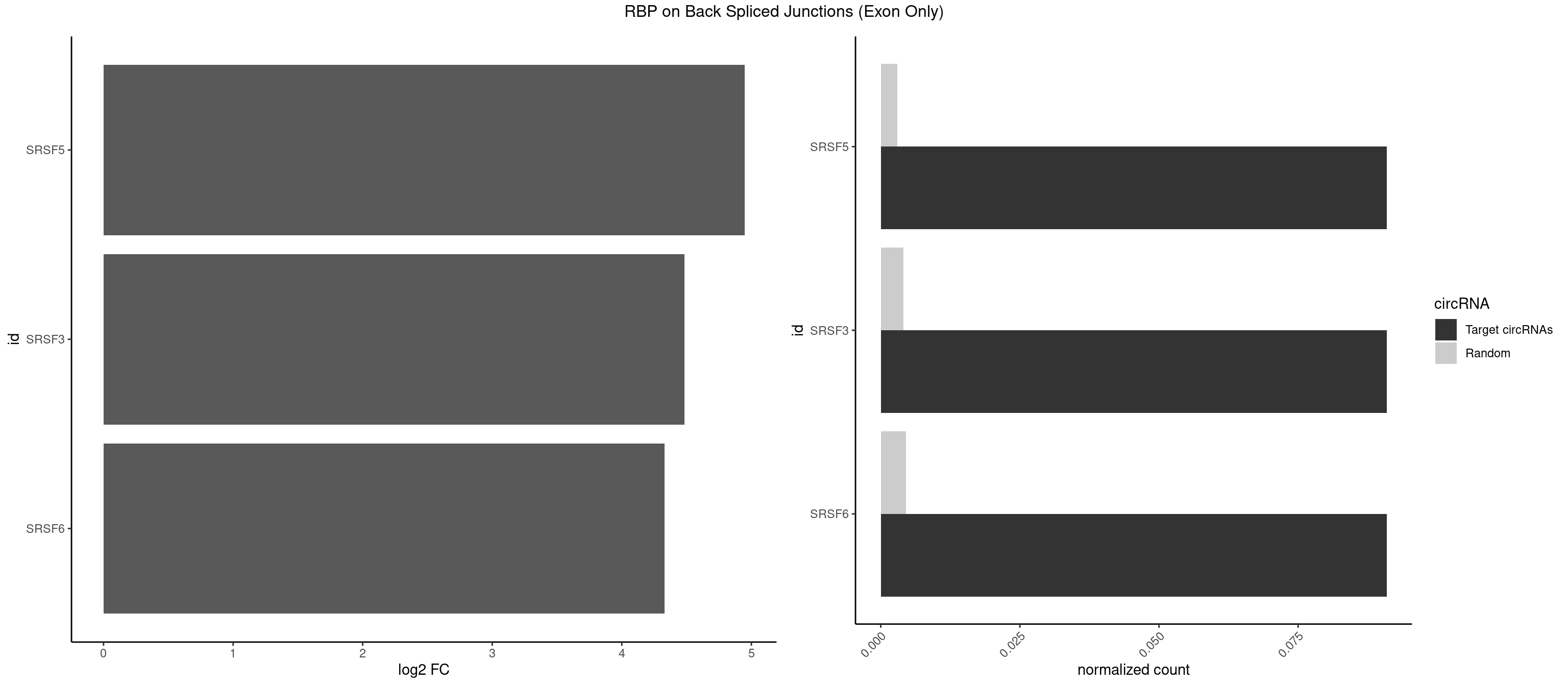
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SRSF5 | 1 | 44 | 0.09090909 | 0.002947341 | 4.946939 | CACAUC | AACAGC,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUC,CACGGA,CGCAGC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACGUA,UGCAGA,UGCAGC,UGCGGC |
| SRSF3 | 1 | 61 | 0.09090909 | 0.004060781 | 4.484596 | CACAUC | AACGAU,ACCACC,ACUACG,AGAGAU,CACAAC,CACAUC,CACCAC,CAGAGA,CAUCAC,CAUCGU,CAUUCA,CCACCA,CCUCGU,CCUCUU,CUACAG,CUCCAA,CUCGUC,CUCUUC,CUUCAA,CUUUAU,GACUUC,GAGAUU,UACAGC,UCAGAG,UCUCCA,UCUUCA,UCUUCC,UGUCAA,UUCGAC,UUCUCC,UUCUUC |
| SRSF6 | 1 | 68 | 0.09090909 | 0.004519256 | 4.330267 | CAUCCU | AACCUG,ACCGUC,ACCUGG,AGAAGA,AGCGGA,AGGAAG,AUCCUG,CAACCU,CACAGG,CACCUG,CACGGA,CACUCG,CACUGG,CAGCAC,CCUGGC,CUACAG,CUCAGG,CUCUGG,CUUCUG,GAAGAA,GAGGAA,GCACCU,GCAGCA,GGAAGA,UACAGG,UACUGG,UCCUGG,UGCGGC,UGUGGA,UUACUG,UUCAGG,UUCUGG,UUUCAG |
RBP on BSJ (Exon and Intron)
Plot
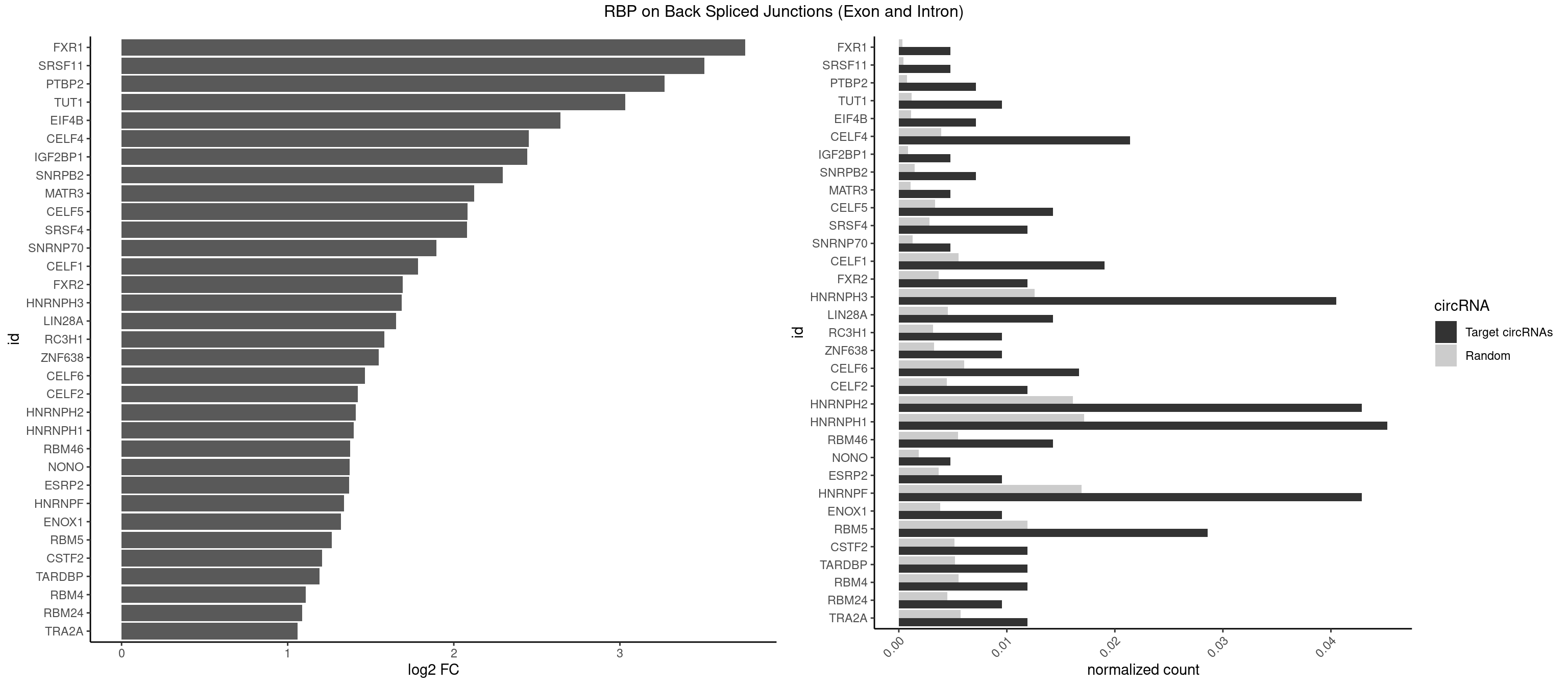
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| FXR1 | 1 | 69 | 0.004761905 | 0.0003526804 | 3.755106 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| SRSF11 | 1 | 82 | 0.004761905 | 0.0004181782 | 3.509349 | AAGAAG | AAGAAG |
| PTBP2 | 2 | 146 | 0.007142857 | 0.0007406288 | 3.269679 | CUCUCU | CUCUCU |
| TUT1 | 3 | 230 | 0.009523810 | 0.0011638452 | 3.032640 | AAAUAC,AAUACU | AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUUGGA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| CELF4 | 8 | 776 | 0.021428571 | 0.0039147521 | 2.452543 | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | CCCGUU | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | GUAUUG,UAUUGC | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AAUCUU | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| CELF5 | 5 | 669 | 0.014285714 | 0.0033756550 | 2.081334 | GUGUGG,GUGUGU,GUGUUG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| SRSF4 | 4 | 558 | 0.011904762 | 0.0028164047 | 2.079612 | AAGAAG,GAAGAA,GAGGAA,GGAAGA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | GAUCAA | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF1 | 7 | 1097 | 0.019047619 | 0.0055320435 | 1.783726 | GUGUGU,GUGUUG,GUUGUG,UGUGUG,UGUUGU,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| FXR2 | 4 | 730 | 0.011904762 | 0.0036829907 | 1.692589 | GACAGG,GGACAG,UGACAG | AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPH3 | 16 | 2496 | 0.040476190 | 0.0125806127 | 1.685871 | AAGGGA,AGGGAA,AGGGGA,GAAGGG,GAGGGG,GGAGGA,GGAGGG,GGGAGG,GGGCUG,GGGGAG,GGGUGU,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| LIN28A | 5 | 900 | 0.014285714 | 0.0045395002 | 1.653968 | GGAGGA,GGAGGG,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| RC3H1 | 3 | 631 | 0.009523810 | 0.0031841999 | 1.580608 | CCUUCU,UCUGUG,UUCUGU | CCCUUC,CCUUCU,CUUCUG,UCCCUU,UCUGUG,UUCUGU |
| ZNF638 | 3 | 646 | 0.009523810 | 0.0032597743 | 1.546767 | GGUUGU,UGUUCU,UGUUGU | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
| CELF6 | 6 | 1196 | 0.016666667 | 0.0060308343 | 1.466536 | GUGUGG,GUGUUG,UGUGGG,UGUGGU,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| CELF2 | 4 | 881 | 0.011904762 | 0.0044437727 | 1.421682 | GUGUGU,UGUGUG,UGUUGU,UUGUGU | AUGUGU,GUAUGU,GUCUGU,GUGUGU,GUUGUU,UAUGUG,UAUGUU,UGUGUG,UGUUGU,UUGUGU |
| HNRNPH2 | 17 | 3198 | 0.042857143 | 0.0161174929 | 1.410908 | AAGGGA,AGGGAA,AGGGGA,GAAGGG,GAGGGG,GGAGGA,GGAGGG,GGGAGG,GGGCUG,GGGGAG,GGGUGU,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 18 | 3406 | 0.045238095 | 0.0171654575 | 1.398030 | AAGGGA,AGGGAA,AGGGGA,GAAGGG,GAGGAA,GAGGGG,GGAGGA,GGAGGG,GGGAGG,GGGCUG,GGGGAG,GGGUGU,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| RBM46 | 5 | 1091 | 0.014285714 | 0.0055018138 | 1.376594 | AUCAAA,AUCAUU,AUGAUA,GAUCAA,GAUGAU | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| ESRP2 | 3 | 731 | 0.009523810 | 0.0036880290 | 1.368689 | GGGAAA,GGGGAG | GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| HNRNPF | 17 | 3360 | 0.042857143 | 0.0169336961 | 1.339639 | AAGGGA,AGGGAA,AGGGGA,GAAGGG,GAGGAA,GAGGGG,GGAGGA,GGAGGG,GGGAGG,GGGCUG,GGGGAG,UGGGGU,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| ENOX1 | 3 | 756 | 0.009523810 | 0.0038139863 | 1.320239 | AGUACA,GGACAG,UGGACA | AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| RBM5 | 11 | 2359 | 0.028571429 | 0.0118903668 | 1.264780 | AGGGAA,AGGGAG,AGGGUG,CAAGGG,GAAGGG,GAGGGA,GAGGGU,UCUUCU | AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUGU,GUGUUG,UGUGUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GAAUGA,GUGUGU,GUUGUG,UGUGUG | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBM4 | 4 | 1094 | 0.011904762 | 0.0055169287 | 1.109602 | CCUUCC,CCUUCU,CUUCUU,UCCUUC | CCUCUU,CCUUCC,CCUUCU,CGCGCG,CGCGGG,CGCGGU,CUCUUU,CUUCCU,CUUCUU,GCGCGA,GCGCGC,GCGCGG,UCCUUC,UUCCUU,UUCUUG |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | GUGUGG,GUGUGU,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| TRA2A | 4 | 1133 | 0.011904762 | 0.0057134220 | 1.059112 | AAGAAA,AAGAAG,GAAGAA,GAGGAA | AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.