circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000180198_ENSG00000180098:+:1:28508127:28553737
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000180198_ENSG00000180098:+:1:28508127:28553737 | ENSG00000180198_ENSG00000180098 | MSTRG.692.8 | + | 1 | 28508128 | 28553737 | 2693 | GAUUUGUUAAGGAUUCCAAGUAACUCUUAUUUGGAGAGAAGACGAUCUGCACUUCGCAUUUUGGCAUUGACAUUUAAUUUUAGGGUCCUUUAUAUAGAAGGGAGACUACAUGAAUGUGUAAGAUCUUGGAGGAAGACAGCAGAGAGAGAGAGAGAGAUCAGAGAUCCCAGGGUUAAAAGUUGGAGAAAUUUCACAGUACAUCAUCCAAAAGAGGAGUCCAUGAUGGAGGCAGAGGUAAACUUGGAGAGACCCUACCGCACAAGAUUUUCCUAAACAGACCGAACUCAAGGAGUCUUUCUGGUUGUUAGUCCACGUGUCCCGAUUUGGGGUUUCCAAAAUACACGCCCACUGGAACCGGGCCAGGGGAGCCAGCCUGGCCAAGGGCUCCCCCAGCCCGGCCAAGGGCUCCCCCAGCCCGGGAGCGCGCCACAUGCAGAUCCUGGGAUGGCCGCCAGGGGCCGCCGGGCUCUUUGUUUUCCUUUCUCACCCGGGUCGGGGCCAGAGGCCUGCAGAGCGCAUGCUCUGGGGCAGUUCGCGGCCCGGCGGGGAGCGCCGGAGUUCCUUGUGGCCGACGUGCACCAAGGACAGGAAGAUGUCACCCAAGCGCAUAGCUAAAAGAAGGUCCCCCCCAGCAGAUGCCAUCCCCAAAAGCAAGAAGGUGAAGGUCUCACACAGGUCCCACAGCACAGAACCCGGCUUGGUGCUGACACUAGGCCAGGGCGACGUGGGCCAGCUGGGGCUGGGUGAGAAUGUGAUGGAGAGGAAGAAGCCGGCCCUGGUAUCCAUUCCGGAGGAUGUUGUGCAGGCUGAGGCUGGGGGCAUGCACACCGUGUGUCUAAGCAAAAGUGGCCAGGUCUAUUCCUUCGGCUGCAAUGAUGAGGGUGCCCUGGGAAGGGACACAUCAGUGGAGGGCUCGGAGAUGGUCCCUGGGAAAGUGGAGCUGCAAGAGAAGGUGGUACAGGUGUCAGCAGGAGACAGUCACACAGCAGCCCUCACCGAUGAUGGCCGUGUCUUCCUCUGGGGCUCCUUCCGGGACAAUAACGGUGUGAUUGGACUGUUGGAGCCCAUGAAGAAGAGCAUGGUGCCUGUGCAGGUGCAGCUGGAUGUGCCUGUGGUAAAGGUGGCCUCAGGAAACGACCACUUGGUGAUGCUGACAGCUGAUGGUGACCUCUACACCUUGGGCUGCGGGGAACAGGGCCAGCUAGGCCGUGUGCCUGAGUUAUUUGCCAACCGUGGUGGCCGGCAAGGCCUCGAACGACUCCUGGUCCCCAAGUGUGUGAUGCUGAAAUCCAGGGGAAGCCGGGGCCACGUGAGAUUCCAGGAUGCCUUUUGUGGUGCCUAUUUCACCUUUGCCAUCUCCCAUGAGGGCCACGUGUACGGCUUCGGCCUCUCCAACUACCAUCAGCUUGGAACUCCGGGCACAGAAUCUUGCUUCAUACCCCAGAACCUAACAUCCUUCAAGAAUUCCACCAAGUCCUGGGUGGGCUUCUCUGGUGGCCAGCACCAUACAGUCUGCAUGGAUUCGGAAGGAAAAGCAUACAGCCUGGGCCGGGCUGAGUAUGGGCGGCUGGGCCUUGGAGAGGGUGCUGAGGAGAAGAGCAUACCCACCCUCAUCUCCAGGCUGCCUGCUGUCUCCUCGGUGGCUUGUGGGGCCUCUGUGGGGUAUGCUGUGACCAAGGAUGGUCGUGUUUUCGCCUGGGGCAUGGGCACCAACUACCAGCUGGGCACAGGGCAGGAUGAGGACGCCUGGAGCCCUGUGGAGAUGAUGGGCAAACAGCUGGAGAACCGUGUGGUCUUAUCUGUGUCCAGCGGGGGCCAGCAUACAGUCUUAUUAGUCAAGGACAAAGAACAGAGCUGAUGAAGCCUCUGAGGGCCUGGCUUCUGUCCUGCACAACCUCCCUCACAGAACAGGGAAGCAGUGACAGCUGCAGAUGGCAGCGGGCCUCUCCCCAGCCCUGAGCACUGUGUCAGUUCCUGCCUUUUCUCAUCAGCAGAACAGAAUCCUUUUCCUCUUUUCCUUCCUCCUCUUUGGAAUUUUCCUGGGACCUACAGAAUAAAGGGGGGGAUGGACAGGGGGUUUUCAAAAGGAACAUGGCUCACUCAGAGCUAUAUGGUUAGACGUUUCUCCCCUUUUCCCUACCUUCCAUGGUCCUGGUUGGCCCUGGCUUUGCCUACUAGAAAACCAAAACUUCCCCCCUGGGGUUUUGUGCCCACUCUCUGAGAAGUUGGGGCUCCAUCAAGCCCCAUUCUAGUCAUGUGCCCCUUUCCUGUCCCUAACAGUCCACAGGCAAACAAAUGGUACAGUCAUAAGAGCCAUCUGUCACGGACCCACGCCCAGAGGAACGUGCAGAAAAAAGCAGAGCUACAUGGCUGUGGGCAACUAUAAGCCAAAUAUUUGGCUCAGAACAGGUGUCCAUGGGACAAAAAAGAACGAUCCUCCACUUGACCAAGAAAAAAGUGAUUCUCCCAGAAGCACAAAGCAUACUCUUGCCCCUCAGGUGUUGCUUGUGUACAUCGUACCCAUCCAUUCGGCUUCACCUGCAGCCAACGGCCUGGAAUCGCAAAGAGACACCACUCUGGGCAGAGCAGAGCAGGCUGGAACCCUACAUGGAUGAGAACUUCAUCUCCAGAGCCUUUGCCACCAUGGGGGAGACCGUAAUGAGCGUCAAAAUUAUCCGAAACCGCCUCACUGGGAUUUGUUAAGGAUUCCAAGUAACUCUUAUUUGGAGAGAAGACGAUCUGC | circ |
| ENSG00000180198_ENSG00000180098:+:1:28508127:28553737 | ENSG00000180198_ENSG00000180098 | MSTRG.692.8 | + | 1 | 28508128 | 28553737 | 22 | CGCCUCACUGGGAUUUGUUAAG | bsj |
| ENSG00000180198_ENSG00000180098:+:1:28508127:28553737 | ENSG00000180198_ENSG00000180098 | MSTRG.692.8 | + | 1 | 28507928 | 28508137 | 210 | UUAAGAAAAAUUGGCUAAAAAGUAGCCAGGCAUGAUGAUAGGUAGCUGGAGGAAGGAGAAUCGCUGGAGCCCAGGAGUGACCUAUACUCAAACCUAUACUCCAGUGCCACUGUACUCCAACCCCAGGCGAUAGCAUGAGGCCCCUCGUUGAAAAAGUUUAGGGUUUUGCUGUACUAAUAGAUUAAUAUCUUGUUUUGCAGGAUUUGUUAA | ie_up |
| ENSG00000180198_ENSG00000180098:+:1:28508127:28553737 | ENSG00000180198_ENSG00000180098 | MSTRG.692.8 | + | 1 | 28553728 | 28553937 | 210 | GCCUCACUGGGUAAGUCUCAUCUCAGGUCUCUCUUAAUACAUCUCGUUGCAGCUGUCAUGUACGAGAAUGUAAACAUCGUAGUCAUGGCUUCUCACUACCCCUAGUAAUAGUCACUGUAAAAUUAUAACAGCUAACAUUUUGAGGUCCCAAUGAGUGGAAGGAUGGCAUGUAGCUUAAGGGUAGGACUUGCCAGGCGUUGUGACUCACAC | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
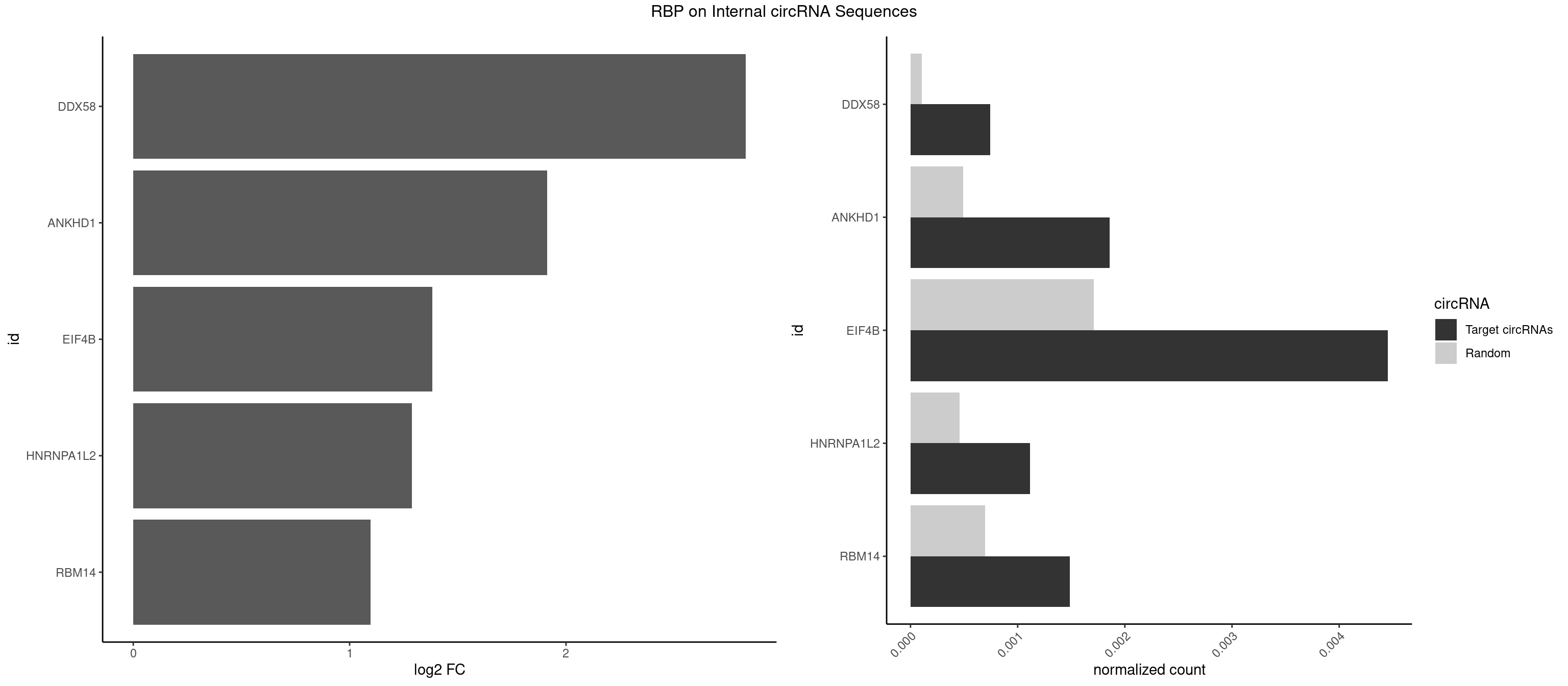
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| DDX58 | 1 | 71 | 0.0007426662 | 0.0001043427 | 2.831384 | GCGCGC | GCGCGC |
| ANKHD1 | 4 | 339 | 0.0018566654 | 0.0004927293 | 1.913847 | AGACGA,AGACGU,GACGAU,GACGUU | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| EIF4B | 11 | 1179 | 0.0044559970 | 0.0017100607 | 1.381701 | CUCGGA,CUUGGA,GUUGGA,UCGGAA,UUGGAA,UUGGAC | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPA1L2 | 2 | 314 | 0.0011139993 | 0.0004564992 | 1.287064 | UAGGGU,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| RBM14 | 3 | 478 | 0.0014853323 | 0.0006941687 | 1.097428 | CGCGCC,CGCGGC,GCGCGC | CGCGCC,CGCGCG,CGCGGC,CGCGGG,GCGCGC,GCGCGG |
RBP on BSJ (Exon Only)
Plot
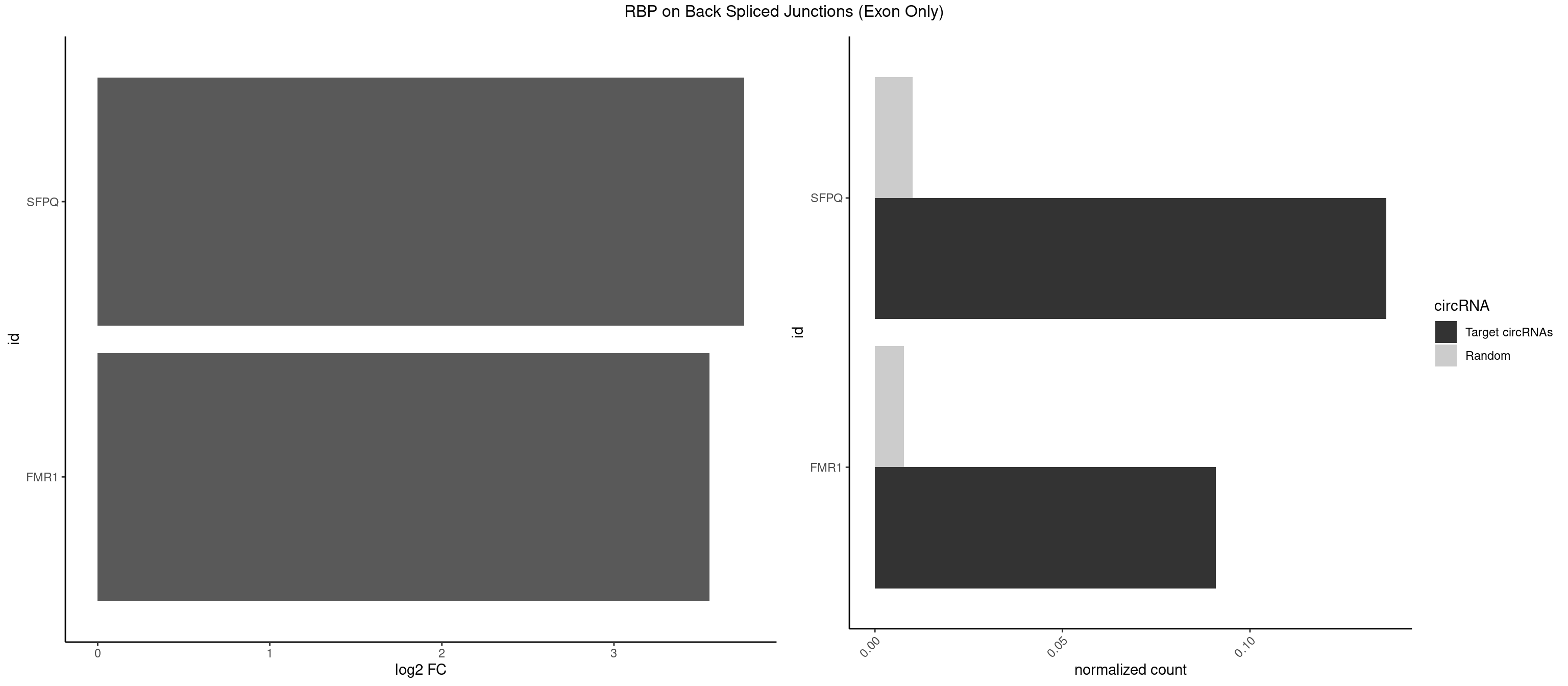
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SFPQ | 2 | 153 | 0.13636364 | 0.010086455 | 3.756968 | ACUGGG,CUGGGA | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
| FMR1 | 1 | 117 | 0.09090909 | 0.007728583 | 3.556149 | ACUGGG | AAAAAA,AAGGAA,AAGGAU,AAGGGA,ACUGGG,ACUGGU,AGCAGC,AGCGAA,AGGAAG,AGGAAU,AGGAGG,AGGAGU,AGGAUG,AGGAUU,AGGCAC,AGGGAG,AGUGGC,AUGGAG,CAGCUG,CUAUGG,CUGAGG,CUGGGC,CUGGUG,GAAGGA,GAAGGG,GACAAG,GACAGG,GAGGAU,GCAGCU,GCAUAG,GCGAAG,GCUAAG,GCUGAG,GCUGGG,GGACAA,GGACAG,GGCAUA,GGCUGA,GGCUGG,GGGCAU,GGGCUA,GGGCUG,GUGCGA,GUGGCU,UAAGGA,UAGUGG,UAUGGA,UGACAA,UGACAG,UGAGGA,UGCGGC,UGGAGU,UGGCUG |
RBP on BSJ (Exon and Intron)
Plot
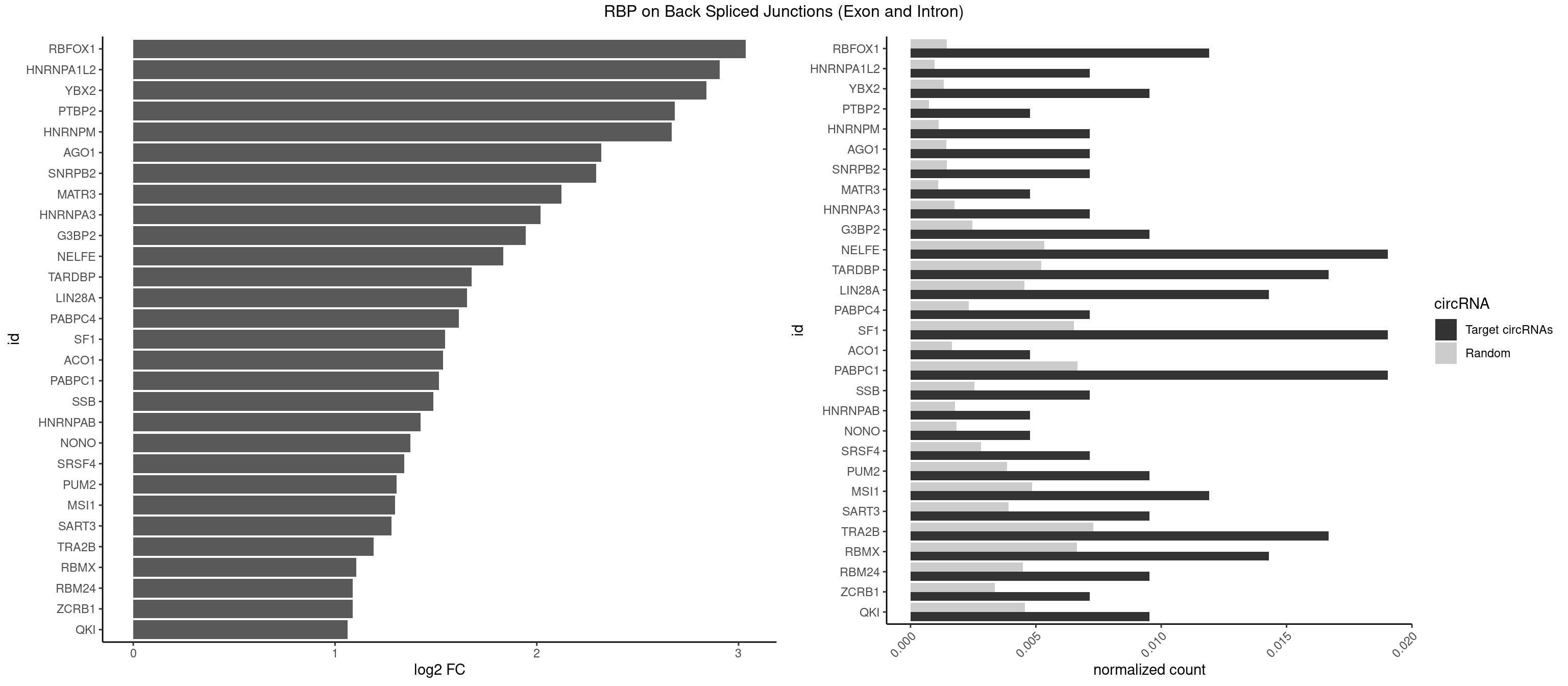
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX1 | 4 | 287 | 0.011904762 | 0.0014510278 | 3.036392 | AGCAUG,GCAUGA,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| HNRNPA1L2 | 2 | 188 | 0.007142857 | 0.0009522370 | 2.907109 | UAGGGU,UUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| YBX2 | 3 | 263 | 0.009523810 | 0.0013301088 | 2.839994 | AACAUC,ACAUCG,ACAUCU | AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| PTBP2 | 1 | 146 | 0.004761905 | 0.0007406288 | 2.684716 | CUCUCU | CUCUCU |
| HNRNPM | 2 | 222 | 0.007142857 | 0.0011235389 | 2.668451 | GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| AGO1 | 2 | 283 | 0.007142857 | 0.0014308746 | 2.319604 | AGGUAG,UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| SNRPB2 | 2 | 288 | 0.007142857 | 0.0014560661 | 2.294425 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| MATR3 | 1 | 216 | 0.004761905 | 0.0010933091 | 2.122837 | AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AAGGAG,GGAGCC | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| G3BP2 | 3 | 490 | 0.009523810 | 0.0024738009 | 1.944809 | AGGAUG,AGGAUU,GGAUGG | AGGAUA,AGGAUG,AGGAUU,GGAUAA,GGAUAG,GGAUGA,GGAUGG,GGAUUA,GGAUUG |
| NELFE | 7 | 1059 | 0.019047619 | 0.0053405885 | 1.834540 | CUCUCU,GCUAAC,GGCUAA,GGUCUC,GUCUCU,UCUCUC,UGGCUA | CUCUCU,CUCUGG,CUGGCU,CUGGUU,GCUAAC,GGCUAA,GGUCUC,GGUUAG,GUCUCU,UCUCUC,UCUCUG,UCUGGC,UCUGGU,UGGCUA,UGGUUA |
| TARDBP | 6 | 1034 | 0.016666667 | 0.0052146312 | 1.676328 | GAAUGU,GUUGUG,GUUUUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| LIN28A | 5 | 900 | 0.014285714 | 0.0045395002 | 1.653968 | AGGAGA,AGGAGU,GGAGAA,GGAGGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAG | AAAAAA,AAAAAG |
| SF1 | 7 | 1294 | 0.019047619 | 0.0065245869 | 1.545652 | ACUAAU,CUAACA,GCUAAC,UACUAA,UAGUAA,UAUACU | ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |
| ACO1 | 1 | 325 | 0.004761905 | 0.0016424829 | 1.535660 | CAGUGC | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| PABPC1 | 7 | 1321 | 0.019047619 | 0.0066606207 | 1.515882 | ACUAAU,AGAAAA,CAAACC,CUAACA,CUAAUA,GAAAAA | AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| SSB | 2 | 505 | 0.007142857 | 0.0025493753 | 1.486358 | UGCUGU,UGUUUU | CUGUUU,GCUGUU,UGCUGU,UGUUUU |
| HNRNPAB | 1 | 351 | 0.004761905 | 0.0017734784 | 1.424957 | AUAGCA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | GAGGAA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| SRSF4 | 2 | 558 | 0.007142857 | 0.0028164047 | 1.342647 | AGGAAG,GAGGAA | AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| PUM2 | 3 | 764 | 0.009523810 | 0.0038542926 | 1.305073 | UACAUC,UGUAAA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| MSI1 | 4 | 961 | 0.011904762 | 0.0048468360 | 1.296424 | AGGAAG,AGGUAG,UAGGUA,UAGUAA | AGGAAG,AGGAGG,AGGUAG,AGGUGG,AGUAAG,AGUAGG,AGUUAG,AGUUGG,UAGGAA,UAGGAG,UAGGUA,UAGGUG,UAGUAA,UAGUAG,UAGUUA,UAGUUG |
| SART3 | 3 | 777 | 0.009523810 | 0.0039197904 | 1.280762 | AGAAAA,GAAAAA | AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| TRA2B | 6 | 1447 | 0.016666667 | 0.0072954454 | 1.191898 | AGGAAG,GAAGGA,GGAAGG,UAAGAA | AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBMX | 5 | 1316 | 0.014285714 | 0.0066354293 | 1.106311 | AGGAAG,GAAGGA,GGAAGG | AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| RBM24 | 3 | 888 | 0.009523810 | 0.0044790407 | 1.088349 | GAGUGA,GAGUGG,UGAGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| ZCRB1 | 2 | 666 | 0.007142857 | 0.0033605401 | 1.087808 | GAUUAA,GCUUAA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| QKI | 3 | 904 | 0.009523810 | 0.0045596534 | 1.062615 | ACUAAU,CUAACA,UACUCA | AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.