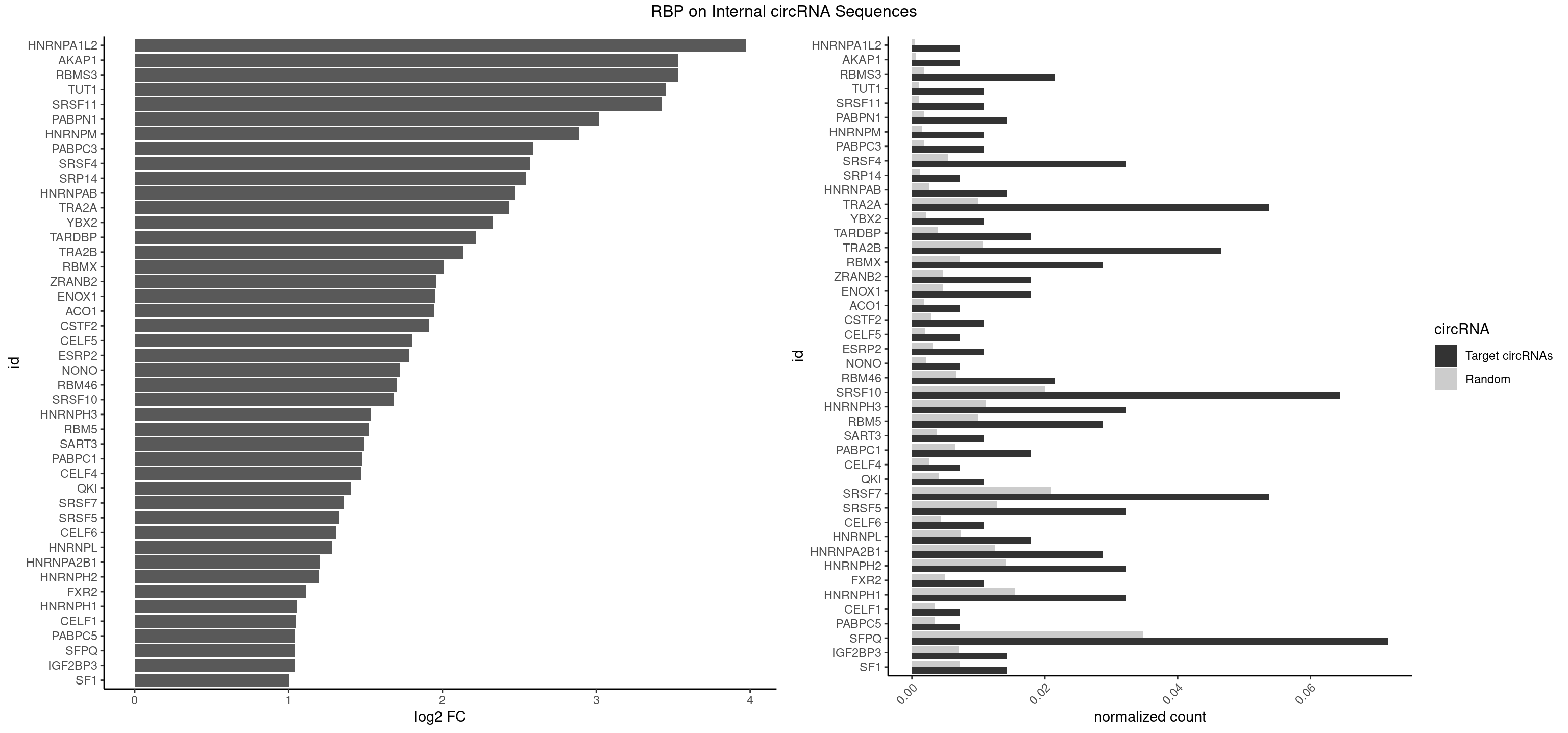
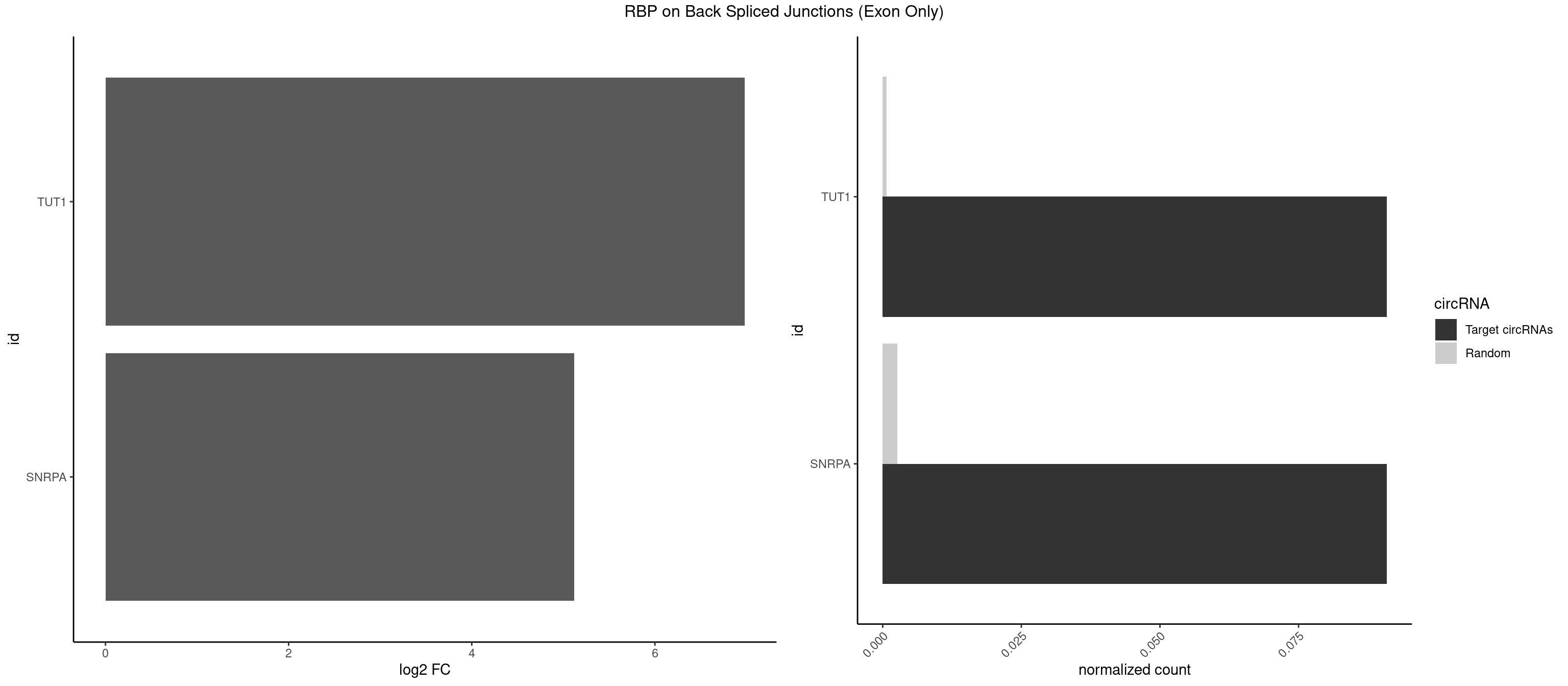
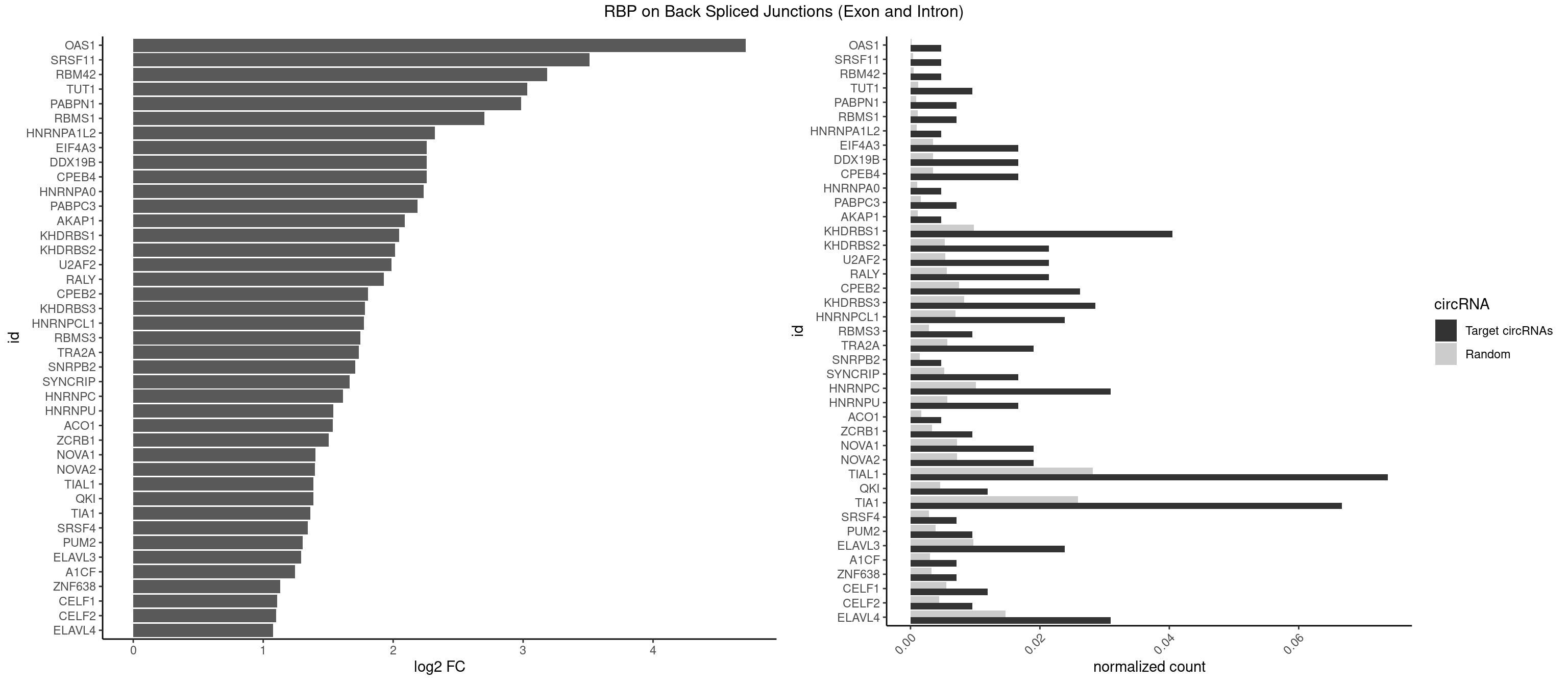
| HNRNPA1L2 |
1 |
314 |
0.007168459 |
0.0004564992 |
3.972979 |
GUAGGG |
AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| AKAP1 |
1 |
426 |
0.007168459 |
0.0006188101 |
3.534094 |
AUAUAU |
AUAUAU,UAUAUA |
| RBMS3 |
5 |
1283 |
0.021505376 |
0.0018607779 |
3.530720 |
AAUAUA,AUAUAU,CAUAUA,CUAUAU,UAUAUC |
AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
| TUT1 |
2 |
678 |
0.010752688 |
0.0009840095 |
3.449881 |
AAAUAC,AGAUAC |
AAAUAC,AAUACU,AGAUAC,CAAUAC,CGAUAC,GAUACU |
| SRSF11 |
2 |
688 |
0.010752688 |
0.0009985015 |
3.428789 |
AAGAAG |
AAGAAG |
| PABPN1 |
3 |
1222 |
0.014336918 |
0.0017723764 |
3.015978 |
AAAAGA,AGAAGA |
AAAAGA,AGAAGA |
| HNRNPM |
2 |
999 |
0.010752688 |
0.0014492040 |
2.891365 |
AAGGAA |
AAGGAA,GAAGGA,GGGGGG |
| PABPC3 |
2 |
1234 |
0.010752688 |
0.0017897669 |
2.586854 |
AAAACA,GAAAAC |
AAAAAC,AAAACA,AAAACC,GAAAAC |
| SRSF4 |
8 |
3740 |
0.032258065 |
0.0054214720 |
2.572903 |
AAGAAG,AGAAGA,GAAGAA,GGAAGA |
AAGAAG,AGAAGA,AGGAAG,GAAGAA,GAGGAA,GGAAGA |
| SRP14 |
1 |
847 |
0.007168459 |
0.0012289250 |
2.544266 |
GCCUGU |
CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| HNRNPAB |
3 |
1782 |
0.014336918 |
0.0025839306 |
2.472096 |
AAGACA,AGACAA,GACAAA |
AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| TRA2A |
14 |
6871 |
0.053763441 |
0.0099589296 |
2.432563 |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,GAAAGA,GAAGAA,GAAGAG |
AAAGAA,AAGAAA,AAGAAG,AAGAGG,AGAAAG,AGAAGA,AGAGGA,AGGAAG,GAAAGA,GAAGAA,GAAGAG,GAGGAA |
| YBX2 |
2 |
1480 |
0.010752688 |
0.0021462711 |
2.324793 |
AACAAC,ACAACA |
AACAAC,AACAUC,ACAACA,ACAACG,ACAACU,ACAUCA,ACAUCG,ACAUCU |
| TARDBP |
4 |
2654 |
0.017921147 |
0.0038476365 |
2.219619 |
GAAUGU,GUGAAU,GUUUUG,UGAAUG |
GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| TRA2B |
12 |
7329 |
0.046594982 |
0.0106226650 |
2.133029 |
AAAGAA,AAGAAG,AAGGAA,AGAAGA,AGGAAA,GAAAGA,GAAGAA |
AAAGAA,AAGAAC,AAGAAG,AAGAAU,AAGGAA,AAUAAG,AGAAGA,AGAAGG,AGGAAA,AGGAAG,AUAAGA,GAAAGA,GAAGAA,GAAGGA,GAAUUA,GGAAGG,UAAGAA |
| RBMX |
7 |
4925 |
0.028673835 |
0.0071387787 |
2.005986 |
AAGAAG,AAGGAA,ACCAAA,AGUGUU,AUCAAA |
AAGAAG,AAGGAA,AAGUAA,AAGUGU,ACCAAA,AGAAGG,AGGAAG,AGUAAC,AGUGUU,AUCAAA,AUCCCA,AUCCCC,GAAGGA,GGAAGG,GUAACA,UAACAA,UAAGAC,UCAAAA |
| ZRANB2 |
4 |
3173 |
0.017921147 |
0.0045997733 |
1.962028 |
AGGGAA,AGGUUU,UGGUGG |
AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,CGGUAA,CGGUAG,CGGUAU,CGGUCU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,GUGGUG,UAAAGG,UGGUAA,UGGUGG |
| ENOX1 |
4 |
3195 |
0.017921147 |
0.0046316558 |
1.952063 |
AAGACA,AAUACA,AGGACA,AUACAG |
AAGACA,AAUACA,AGACAG,AGGACA,AGUACA,AUACAG,CAGACA,CAUACA,CGGACA,CGUACA,GGACAG,GUACAG,UAGACA,UAUACA,UGGACA,UGUACA |
| ACO1 |
1 |
1283 |
0.007168459 |
0.0018607779 |
1.945757 |
CAGUGU |
CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| CSTF2 |
2 |
1967 |
0.010752688 |
0.0028520334 |
1.914635 |
GUGUUG,GUUUUG |
GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| CELF5 |
1 |
1415 |
0.007168459 |
0.0020520728 |
1.804581 |
GUGUUG |
GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| ESRP2 |
2 |
2150 |
0.010752688 |
0.0031172377 |
1.786357 |
GGGAAA,GGGAAG |
GGGAAA,GGGAAG,GGGAAU,GGGGAA,GGGGAG,GGGGAU,UGGGAA,UGGGGA |
| NONO |
1 |
1498 |
0.007168459 |
0.0021723567 |
1.722402 |
AGGAAC |
AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBM46 |
5 |
4554 |
0.021505376 |
0.0066011240 |
1.703914 |
AUCAAA,AUCAUG,AUGAAG,GAUGAA |
AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| SRSF10 |
17 |
13860 |
0.064516129 |
0.0200874160 |
1.683368 |
AAAAGA,AAAGAA,AAAGGA,AAGAAA,AAGAAG,AAGACA,AAGAGG,AAGGAA,AGACAA,AGAGAA,AGAGGG,GAAAGA,GACAAA,GAGAAG |
AAAAGA,AAAGAA,AAAGAC,AAAGAG,AAAGGA,AAAGGG,AAGAAA,AAGAAG,AAGACA,AAGAGA,AAGAGG,AAGGAA,AAGGAG,AAGGGA,AAGGGG,ACAAAG,AGACAA,AGAGAA,AGAGAC,AGAGAG,AGAGGA,AGAGGG,CAAAGA,GAAAGA,GACAAA,GAGAAA,GAGAAC,GAGAAG,GAGACA,GAGACC,GAGACG,GAGAGA,GAGAGC,GAGAGG,GAGGAA,GAGGAG,GAGGGA,GAGGGG |
| HNRNPH3 |
8 |
7688 |
0.032258065 |
0.0111429292 |
1.533531 |
AAUGUG,AGGGAA,AUGUGG,GAAUGU,GGGAAG,UCGAGG |
AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| RBM5 |
7 |
6879 |
0.028673835 |
0.0099705232 |
1.523994 |
AAGGAA,AGGGAA,AGGGUA,GAGGGU,UUCUCU |
AAAAAA,AAGGAA,AAGGAG,AAGGGG,AAGGUA,AAGGUG,AGGGAA,AGGGAG,AGGGUA,AGGGUG,AGGUAA,CAAGGA,CAAGGG,CCCCCC,CUCUUC,CUUCUC,GAAGGA,GAAGGG,GAAGGU,GAGGGA,GAGGGU,GGGGGG,GGUGGU,UCUUCU,UUCUCU |
| SART3 |
2 |
2634 |
0.010752688 |
0.0038186524 |
1.493562 |
GAAAAA,GAAAAC |
AAAAAA,AAAAAC,AGAAAA,GAAAAA,GAAAAC |
| PABPC1 |
4 |
4443 |
0.017921147 |
0.0064402624 |
1.476472 |
CAAAUA,CAAAUC,GAAAAA,GAAAAC |
AAAAAA,AAAAAC,ACAAAC,ACAAAU,ACGAAC,ACGAAU,ACUAAC,ACUAAU,AGAAAA,CAAACA,CAAACC,CAAAUA,CAAAUC,CGAACA,CGAACC,CGAAUA,CGAAUC,CUAACA,CUAACC,CUAAUA,CUAAUC,GAAAAA,GAAAAC |
| CELF4 |
1 |
1782 |
0.007168459 |
0.0025839306 |
1.472096 |
GUGUUG |
GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| QKI |
2 |
2805 |
0.010752688 |
0.0040664663 |
1.402850 |
AUUAAC,UCUACU |
AAUCAU,ACACAC,ACACUA,ACUAAC,ACUAAU,ACUCAU,ACUUAU,AUCAUA,AUCUAA,AUCUAC,AUUAAC,CACACU,CACUAA,CUAACA,CUAACC,CUAACG,CUAAUC,CUACUC,CUCAUA,UAACCU,UAAUCA,UACUCA,UCAUAU,UCUAAU,UCUACU |
| SRSF7 |
14 |
14481 |
0.053763441 |
0.0209873716 |
1.357104 |
AAAGGA,AAGAAG,AGAAGA,AGAGAA,AGGACA,AUUGAC,CAGAGA,CUUCAC,GAAGAA,GACAAA |
AAAGGA,AAGAAG,AAGGAC,AAGGCG,AAUGAU,ACGAAU,ACGACG,ACGAGA,ACGAUA,ACGAUU,ACUACG,ACUAGA,AGAAGA,AGACGA,AGACUA,AGAGAA,AGAGAC,AGAGAG,AGAGAU,AGAGGA,AGAUCA,AGAUCU,AGGAAG,AGGACA,AGGCGA,AUAGAA,AUAGAC,AUUGAC,AUUGAU,CAGAGA,CCGAGA,CGAAUG,CGACGA,CGAGAC,CGAGAG,CGAGAU,CGAUAG,CGAUUG,CUACGA,CUAGAG,CUCUUC,CUGAGA,CUUCAC,GAAGAA,GAAGGC,GAAUGA,GACAAA,GACGAC,GACGAG,GACGAU,GACUAC,GAGACU,GAGAGA,GAGAUC,GAGGAA,GAUAGA,GAUUGA,GGAAGG,GGACAA,GGACGA,UACGAC,UACGAU,UAGAGA,UCAACA,UCGAGA,UCGAUU,UCUCCG,UCUUCA,UGAGAG,UGGACA |
| SRSF5 |
8 |
8869 |
0.032258065 |
0.0128544391 |
1.327393 |
AAGAAG,AGAAGA,GAAGAA,GGAAGA |
AACAGC,AAGAAG,AGAAGA,AGGAAG,AUAAAG,CAACAG,CACAGA,CACAGC,CACAUA,CACAUC,CACGGA,CACGGC,CACGUA,CACGUC,CCGCAG,CGCAGA,CGCAGC,CGCAGG,CGCAUA,CGCAUC,CGCGGA,CGCGGC,CGCGUA,CGCGUC,GAAGAA,GAGGAA,GGAAGA,UAAAGG,UACAGA,UACAGC,UACAUA,UACAUC,UACGGA,UACGGC,UACGUA,UACGUC,UGCAGA,UGCAGC,UGCAUA,UGCAUC,UGCGGA,UGCGGC,UGCGUA,UGCGUC |
| CELF6 |
2 |
2997 |
0.010752688 |
0.0043447134 |
1.307364 |
GUGUUG,UGUGAU |
GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| HNRNPL |
4 |
5085 |
0.017921147 |
0.0073706513 |
1.281799 |
AAACAA,AAAUAC,AAUACA,ACAUAA |
AAACAA,AAACAC,AAAUAA,AAAUAC,AACAAA,AACACA,AAUAAA,AAUACA,ACACAA,ACACAC,ACACCA,ACACGA,ACAUAA,ACAUAC,ACCACA,CACAAA,CACAAC,CACAAG,CACACA,CACACC,CACCAC,CACGAA,CACGAC,CACGAG,CAUAAA,CAUACA |
| HNRNPA2B1 |
7 |
8607 |
0.028673835 |
0.0124747476 |
1.200724 |
AAGAAG,AAGGAA,AGAAGC,AGGAAC,GUAGGG |
AAGAAG,AAGGAA,AAGGAG,AAGGGG,AAUUUA,ACUAGA,AGAAGC,AGACUA,AGAUAU,AGGAAC,AGGAGC,AGGGGC,AGUAGG,AUAGCA,AUAGGG,AUUUAA,CAAGAA,CAAGGA,CAAGGG,CCAAGA,CCAAGG,CGAAGG,CUAGAC,GAAGCC,GAAGGA,GACUAG,GCCAAG,GCGAAG,GGAACC,GGAGCC,GGGGCC,GUAGGG,UAGACA,UAGACU,UAGGGA,UAGGGU,UUAGGG,UUUAUA |
| HNRNPH2 |
8 |
9711 |
0.032258065 |
0.0140746688 |
1.196559 |
AAUGUG,AGGGAA,AUGUGG,GAAUGU,GGGAAG,UCGAGG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| FXR2 |
2 |
3434 |
0.010752688 |
0.0049780156 |
1.111055 |
AGACAA,GACAAA |
AGACAA,AGACAG,AGACGA,AGACGG,GACAAA,GACAAG,GACAGA,GACAGG,GACGAA,GACGAG,GACGGA,GACGGG,GGACAA,GGACAG,GGACGA,GGACGG,UGACAA,UGACAG,UGACGA,UGACGG |
| HNRNPH1 |
8 |
10717 |
0.032258065 |
0.0155325680 |
1.054364 |
AAUGUG,AGGGAA,AUGUGG,GAAUGU,GGGAAG,UCGAGG |
AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| CELF1 |
1 |
2391 |
0.007168459 |
0.0034664959 |
1.048185 |
GUGUUG |
CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| PABPC5 |
1 |
2400 |
0.007168459 |
0.0034795387 |
1.042767 |
AGAAAG |
AGAAAA,AGAAAG,AGAAAU,GAAAAU,GAAAGU,GAAAUU |
| SFPQ |
19 |
24050 |
0.071684588 |
0.0348548043 |
1.040305 |
AAGAGG,AAGCAA,AAGGAA,ACUGGG,AGGAAC,GAAGAA,GAAGAG,GAAGCA,GAGGAC,GGAAGA,UAAGGA,UGCAGG,UGGUGG,UUAGUU,UUGAUU,UUGGUG |
AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGAUCG,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUAAGG,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GACUGG,GAGAGG,GAGGAA,GAGGAC,GAGGUA,GAUCGG,GCAGGC,GGAAGA,GGACUG,GGAGAG,GGAGGA,GGAGGG,GGGAUC,GGGGAC,GGGGAU,GGGGGA,GGGGGG,GGUAAG,GGUCUG,GUAAGA,GUAAUG,GUAAUU,GUAGUG,GUAGUU,GUCUGG,GUGAUG,GUGAUU,GUGGUG,GUGGUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUGU,UAAUUG,UAAUUU,UAGAGA,UAGAUC,UAGGGG,UAGUGG,UAGUGU,UAGUUG,UAGUUU,UCGGAA,UCUAAG,UCUGGA,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGAUUU,UGCAGG,UGGAAG,UGGAGA,UGGAGC,UGGAGG,UGGUCU,UGGUGG,UGGUGU,UGGUUG,UGGUUU,UUAAUG,UUAAUU,UUAGUG,UUAGUU,UUGAAG,UUGAUG,UUGAUU,UUGGUG,UUGGUU,UUUUUU |
| IGF2BP3 |
3 |
4815 |
0.014336918 |
0.0069793662 |
1.038567 |
AAAACA,AAAUAC,AAUACA |
AAAAAC,AAAACA,AAAAUC,AAACAC,AAACUC,AAAUAC,AAAUCA,AAAUUC,AACACA,AACUCA,AAUACA,AAUUCA,ACAAAC,ACAAUC,ACACAC,ACACUC,ACAUAC,ACAUUC,CAAACA,CAAUCA,CACACA,CACUCA,CAUACA,CAUUCA |
| SF1 |
3 |
4931 |
0.014336918 |
0.0071474739 |
1.004230 |
AUUAAC,GCUGAC,UGCUGC |
ACAGAC,ACAGUC,ACCGAC,ACGAAC,ACUAAC,ACUAAG,ACUAAU,ACUAGC,ACUGAC,ACUUAU,AGUAAC,AGUAAG,AUACUA,AUUAAC,CACAGA,CACCGA,CACUGA,CAGUCA,CGCUGA,CUAACA,GACUAA,GCUAAC,GCUGAC,GCUGCC,UAACAA,UACGAA,UACUAA,UACUAG,UACUGA,UAGUAA,UAUACU,UGCUAA,UGCUGA,UGCUGC |