circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000141298:-:17:29648143:29672129
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000141298:-:17:29648143:29672129 | ENSG00000141298 | ENST00000540801 | - | 17 | 29648144 | 29672129 | 813 | GUCUGCACUACAGAGCUUACACAAGGCUUGUGAAGUCGCCAGAGCGCAUAACUACUACCCAGGCAGCCUAUUUCUCACUUGGGUGAGUUAUUAUGAGAGCCAUAUCAACUCAGAUCAAUCCUCAGUCAAUGAAUGGAAUGCAAUGCAAGAUGUACAGUCCCACCGGCCCGACUCUCCAGCUCUCUUCACCGACAUACCUACUGAACGUGAACGAACAGAAAGGCUAAUUAAAACCAAAUUAAGGGAGAUCAUGAUGCAGAAGGAUUUGGAGAAUAUUACAUCCAAAGAGAUAAGAACAGAGUUGGAAAUGCAAAUGGUGUGCAACUUGCGGGAAUUCAAGGAAUUUAUAGACAAUGAAAUGAUAGUGAUCCUUGGUCAAAUGGAUAGCCCUACACAGAUAUUUGAGCAUGUGUUCCUGGGCUCAGAAUGGAAUGCCUCCAACUUAGAGGACUUACAGAACCGAGGGGUACGGUAUAUCUUGAAUGUCACUCGAGAGAUAGAUAACUUCUUCCCAGGAGUCUUUGAGUAUCAUAACAUUCGGGUAUAUGAUGAAGAGGCAACGGAUCUCCUGGCGUACUGGAAUGACACUUACAAAUUCAUCUCUAAAGCAAAGAAACAUGGAUCUAAAUGCCUUGUGCACUGCAAAAUGGGGGUGAGUCGCUCAGCCUCCACCGUGAUUGCCUAUGCAAUGAAGGAAUAUGGCUGGAAUCUGGACCGAGCCUAUGACUAUGUGAAAGAAAGACGAACGGUAACCAAGCCCAACCCAAGCUUCAUGAGACAACUGGAAGAGUAUCAGGGGAUCUUGCUGGCAAGGUCUGCACUACAGAGCUUACACAAGGCUUGUGAAGUCGCCAGAGCGCAUA | circ |
| ENSG00000141298:-:17:29648143:29672129 | ENSG00000141298 | ENST00000540801 | - | 17 | 29648144 | 29672129 | 22 | UUGCUGGCAAGGUCUGCACUAC | bsj |
| ENSG00000141298:-:17:29648143:29672129 | ENSG00000141298 | ENST00000540801 | - | 17 | 29672120 | 29672329 | 210 | CUUUCAGCAAAUGUCUUGUCCAUUGCUGUGGAUAGAUAAUAUGGUAUAAUCAGAAUUGGAGAAUCUUGACUUUCAAGGAGUAGAUUGUCCAGAGGACAAAUUCUAUAGAAUUUAAAAAUUAGAAGAGUUUGGAGUACACAGGUUAUUGGCCUUUGGCACCCCACAGUUCUAUGGUGCUCAUUUCCCAGCAUGUCUUUCAGGUCUGCACUA | ie_up |
| ENSG00000141298:-:17:29648143:29672129 | ENSG00000141298 | ENST00000540801 | - | 17 | 29647944 | 29648153 | 210 | UGCUGGCAAGGUGAGUCAGUGACUUUCUCCAAUUCUCCCUCCUUUUAAGUUCCCUAAAGCAAGAUGAAGUGUCCUUCUCUGUAGCUACUUCUCAAGCUGAUUUGGAUGUGAUAUUUUUUCAAAAAAGACUUAUUUCUCUAUCAAUCCAGUUUUCAAAACAUAAGUGUAAGCAUUUAAGAUUUGCAUGGGCUGGCCGGGCACAGUGGCUCA | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
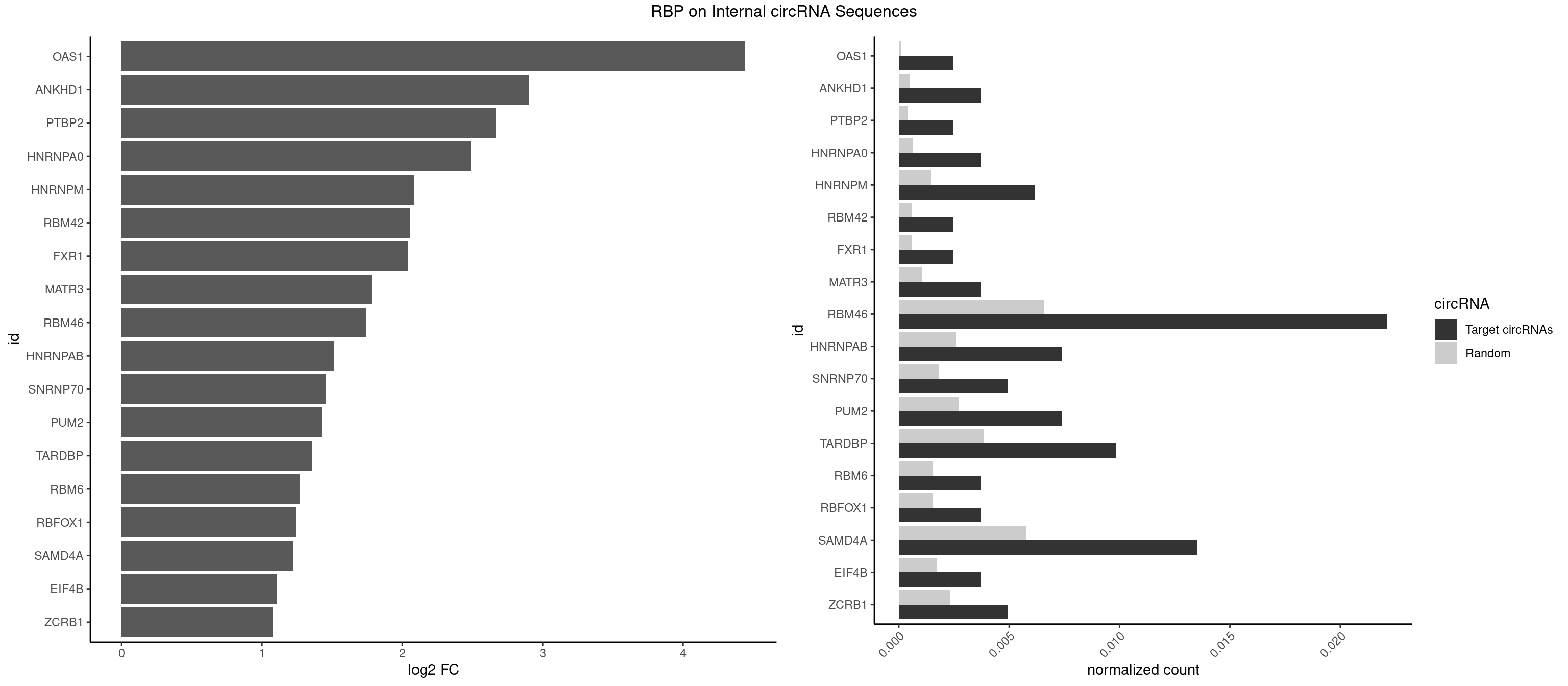
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| OAS1 | 1 | 77 | 0.002460025 | 0.0001130379 | 4.443794 | GCAUAA | GCAUAA |
| ANKHD1 | 2 | 339 | 0.003690037 | 0.0004927293 | 2.904768 | AGACGA,GACGAA | AGACGA,AGACGU,GACGAA,GACGAU,GACGUA,GACGUU |
| PTBP2 | 1 | 267 | 0.002460025 | 0.0003883867 | 2.663107 | CUCUCU | CUCUCU |
| HNRNPA0 | 2 | 453 | 0.003690037 | 0.0006579386 | 2.487610 | AAUUUA,AGAUAU | AAUUUA,AGAUAU,AGUAGG |
| HNRNPM | 4 | 999 | 0.006150062 | 0.0014492040 | 2.085340 | AAGGAA,GAAGGA | AAGGAA,GAAGGA,GGGGGG |
| RBM42 | 1 | 407 | 0.002460025 | 0.0005912752 | 2.056771 | AACUAC | AACUAA,AACUAC,ACUAAG,ACUACG |
| FXR1 | 1 | 411 | 0.002460025 | 0.0005970720 | 2.042696 | AUGACA | ACGACA,ACGACG,AUGACA,AUGACG |
| MATR3 | 2 | 739 | 0.003690037 | 0.0010724109 | 1.782777 | AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| RBM46 | 17 | 4554 | 0.022140221 | 0.0066011240 | 1.745886 | AAUGAA,AAUGAU,AUCAAU,AUCAUA,AUCAUG,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,GAUCAA,GAUCAU,GAUGAA | AAUCAA,AAUCAU,AAUGAA,AAUGAU,AUCAAA,AUCAAG,AUCAAU,AUCAUA,AUCAUG,AUCAUU,AUGAAA,AUGAAG,AUGAAU,AUGAUA,AUGAUG,AUGAUU,GAUCAA,GAUCAU,GAUGAA,GAUGAU |
| HNRNPAB | 5 | 1782 | 0.007380074 | 0.0025839306 | 1.514068 | AAAGAC,AGACAA,CAAAGA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SNRNP70 | 3 | 1237 | 0.004920049 | 0.0017941145 | 1.455401 | AUUCAA,GAUCAA,UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| PUM2 | 5 | 1890 | 0.007380074 | 0.0027404447 | 1.429225 | GUAUAU,UACAUC,UAGAUA,UGUACA | GUAAAU,GUACAU,GUAGAU,GUAUAU,UAAAUA,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUACA,UGUAGA,UGUAUA |
| TARDBP | 7 | 2654 | 0.009840098 | 0.0038476365 | 1.354700 | GAAUGA,GAAUGG,GAAUGU,UGAAUG,UUGUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| RBM6 | 2 | 1054 | 0.003690037 | 0.0015289102 | 1.271132 | AUCCAA,CAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBFOX1 | 2 | 1077 | 0.003690037 | 0.0015622419 | 1.240017 | AGCAUG,GCAUGU | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| SAMD4A | 10 | 3992 | 0.013530135 | 0.0057866714 | 1.225371 | CGGGAA,CGGGUA,CUGGAA,CUGGAC,CUGGCA,GCGGGA,GCUGGA,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| EIF4B | 2 | 1179 | 0.003690037 | 0.0017100607 | 1.109588 | GUUGGA,UUGGAA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| ZCRB1 | 3 | 1605 | 0.004920049 | 0.0023274215 | 1.079940 | AAUUAA,GACUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
RBP on BSJ (Exon Only)
Plot

Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| YBX1 | 1 | 19 | 0.09090909 | 0.001309929 | 6.116864 | GGUCUG | AACCAC,ACACCA,ACAUCU,ACCACC,CACCAC,CAGCAA,CCACCA,CCAGCA,CCUGCG,CUGCGG,GAUCUG,GCCUGC,UCCAGC,UGCGGU |
| ZRANB2 | 1 | 45 | 0.09090909 | 0.003012837 | 4.915230 | AGGUCU | AAAGGU,AGGGAA,AGGUAA,AGGUAC,AGGUAG,AGGUAU,AGGUCU,AGGUUA,AGGUUU,GAGGUU,GGGUAA,GGGUGA,GGUAAA,GGUGGU,GUAAAG,UAAAGG,UGGUAA |
| SFPQ | 1 | 153 | 0.09090909 | 0.010086455 | 3.172005 | GGUCUG | AAGAAC,AAGAGC,AAGAGG,AAGCAA,AAGGAA,AAGGAC,AAGGGA,ACUGGG,AGAGAG,AGAGGA,AGAGGU,AGGAAC,AGGACC,AGGGAU,AGGGGG,AUCGGA,CAGGCA,CUGGAG,CUGGGA,GAAGAA,GAAGAG,GAAGCA,GAAGGA,GAGGAA,GAGGAC,GAGGUA,GCAGGC,GGAAGA,GGAGAG,GGAGGA,GGAGGG,GGGGGA,GUAAGA,GUAAUG,GUAGUG,GUAGUU,GUCUGG,GUGAUU,UAAGAG,UAAGGA,UAAGGG,UAAUGG,UAAUUG,UAGAGA,UAGAUC,UAGUGG,UAGUGU,UAGUUG,UCGGAA,UCUAAG,UGAAGC,UGAUGG,UGAUGU,UGAUUG,UGCAGG,UGGAGA,UGGAGC,UGGAGG,UGGUUU,UUAAUG,UUAGUG,UUAGUU,UUGAAG,UUGGUU |
RBP on BSJ (Exon and Intron)
Plot
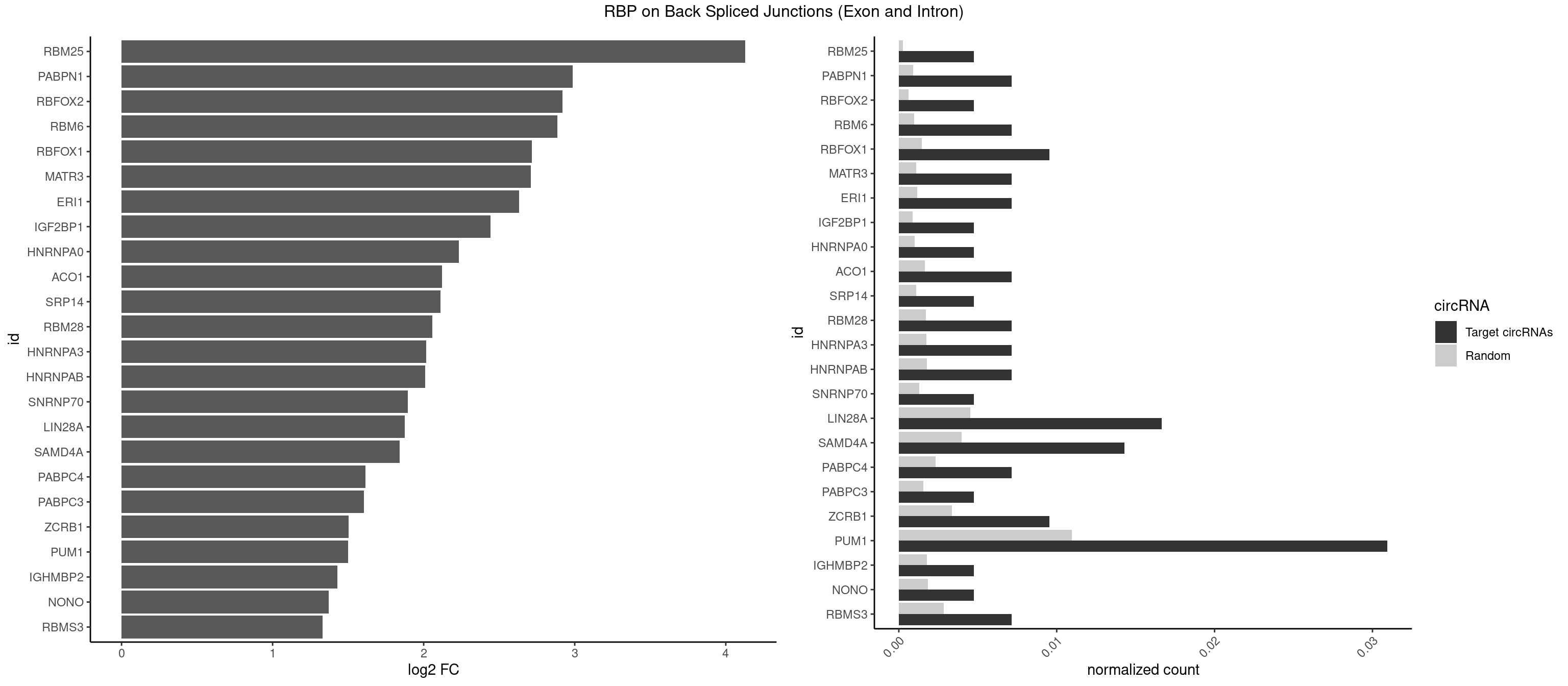
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBM25 | 1 | 53 | 0.004761905 | 0.0002720677 | 4.129501 | CGGGCA | AUCGGG,CGGGCA,UCGGGC |
| PABPN1 | 2 | 178 | 0.007142857 | 0.0009018541 | 2.985535 | AAAAGA,AGAAGA | AAAAGA,AGAAGA |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| RBM6 | 2 | 191 | 0.007142857 | 0.0009673519 | 2.884389 | AAUCCA,AUCCAG | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| RBFOX1 | 3 | 287 | 0.009523810 | 0.0014510278 | 2.714464 | AGCAUG,GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| MATR3 | 2 | 216 | 0.007142857 | 0.0010933091 | 2.707800 | AAUCUU,AUCUUG | AAUCUU,AUCUUA,AUCUUG,CAUCUU |
| ERI1 | 2 | 228 | 0.007142857 | 0.0011537686 | 2.630147 | UUUCAG | UUCAGA,UUUCAG |
| IGF2BP1 | 1 | 173 | 0.004761905 | 0.0008766626 | 2.441445 | GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| HNRNPA0 | 1 | 200 | 0.004761905 | 0.0010126965 | 2.233337 | AAUUUA | AAUUUA,AGAUAU,AGUAGG |
| ACO1 | 2 | 325 | 0.007142857 | 0.0016424829 | 2.120623 | CAGUGA,CAGUGG | CAGUGA,CAGUGC,CAGUGG,CAGUGU |
| SRP14 | 1 | 218 | 0.004761905 | 0.0011033857 | 2.109602 | CUGUAG | CCUGUA,CGCCUG,CUGUAG,GCCUGU |
| RBM28 | 2 | 340 | 0.007142857 | 0.0017180572 | 2.055723 | AGUAGA,GAGUAG | AGUAGA,AGUAGG,AGUAGU,GAGUAG,GUGUAG,UGUAGA,UGUAGG,UGUAGU |
| HNRNPA3 | 2 | 349 | 0.007142857 | 0.0017634019 | 2.018140 | AAGGAG,CAAGGA | AAGGAG,AGGAGC,CAAGGA,CCAAGG,GCCAAG,GGAGCC |
| HNRNPAB | 2 | 351 | 0.007142857 | 0.0017734784 | 2.009919 | AAAGAC,GACAAA | AAAGAC,AAGACA,ACAAAG,AGACAA,AUAGCA,CAAAGA,GACAAA |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| LIN28A | 6 | 900 | 0.016666667 | 0.0045395002 | 1.876360 | AGGAGU,GGAGAA,GGAGUA,UGGAGA,UGGAGU | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| SAMD4A | 5 | 790 | 0.014285714 | 0.0039852882 | 1.841817 | CGGGCA,CUGGCA,CUGGCC,GCUGGC | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| PABPC4 | 2 | 462 | 0.007142857 | 0.0023327287 | 1.614483 | AAAAAA,AAAAAG | AAAAAA,AAAAAG |
| PABPC3 | 1 | 310 | 0.004761905 | 0.0015669085 | 1.603618 | AAAACA | AAAAAC,AAAACA,AAAACC,GAAAAC |
| ZCRB1 | 3 | 666 | 0.009523810 | 0.0033605401 | 1.502846 | AUUUAA,GACUUA | AAUUAA,ACUUAA,AUUUAA,GAAUUA,GACUUA,GAUUAA,GAUUUA,GCUUAA,GGAUUA,GGCUUA,GGUUUA,GUUUAA |
| PUM1 | 12 | 2172 | 0.030952381 | 0.0109482064 | 1.499356 | ACAUAA,AGAAUU,AGAUAA,CAGAAU,GAAUUG,GUAGAU,GUCCAG,UAAUAU,UAGAUA,UGUCCA | AAUAUU,AAUGUU,AAUUGU,ACAUAA,AGAAUU,AGAUAA,AUUGUA,CAGAAU,CCAGAA,CUUGUA,GAAUUG,GUAAAU,GUAAUA,GUACAU,GUAGAU,GUAUAU,GUCCAG,UAAAUA,UAAUAU,UAAUGU,UACAUA,UACAUC,UAGAUA,UAUAUA,UGUAAA,UGUAAU,UGUACA,UGUAGA,UGUAUA,UGUCCA,UUAAUG,UUGUAC,UUGUAG,UUUAAU |
| IGHMBP2 | 1 | 350 | 0.004761905 | 0.0017684401 | 1.429061 | AAAAAA | AAAAAA |
| NONO | 1 | 364 | 0.004761905 | 0.0018389762 | 1.372636 | AGAGGA | AGAGGA,AGGAAC,GAGAGG,GAGGAA |
| RBMS3 | 2 | 562 | 0.007142857 | 0.0028365578 | 1.332360 | CUAUAG,UAUAGA | AAUAUA,AUAUAG,AUAUAU,CAUAUA,CUAUAG,CUAUAU,UAUAGA,UAUAGC,UAUAUA,UAUAUC |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.