circRNA Analysis
WANG Ziyi
Date: 18 5月 2022
ENSG00000156860:+:16:30664214:30666541
1. Sequences
| id | gene | transcript | strand | chrom | startGR | endGR | length | seq | type |
|---|---|---|---|---|---|---|---|---|---|
| ENSG00000156860:+:16:30664214:30666541 | ENSG00000156860 | ENST00000356166 | + | 16 | 30664215 | 30666541 | 748 | CUCUUCACGGCCGCCCCCCAAGGCCCCGGCCCCUCCCGUGGCUCAGCCUCCCCCCUCAUCAUCCUCUUCGUCCUCCUCCUCCUCAUCUGCCUCCUCCUCGUCCGCGCAGCUCACCCACCGGCCCCCGACGCCCUCACUGCCCCUGCCUUUGUCCACCCACAGCUUUCCCCCUCCCGGGCUGCGGCCCCCCCCACCACCCCACCACCCCUCCUUGUUCUCCCCUGGCCCCACCCUGCCCCCACCCCCACCCCUGCUGCAGGUGCCAGGGCACCCUGGGGCCUCAGCCGCUAACGCCCUUUCUGAGCAGGACCUGAUCGGCCAGGACCUGAACUCUCGCUACCUGAAUGCCCAGGGUGGCCCUGAGGUGGUGGGGGCAGGGGGCUCGGCCCGGCCCCUGGCCUUCCAGUUCCACCAGCACAACCACCAGCACCAGCACACCCACCAGCACACCCACCAGCACUUCACCCCUUAUCCCCCGGGCCUGCUGCCACCCCACGGCCCCCACAUGUUUGAGAAAUAUCCAGGAAAGAUGGAAGGCCUUUUCCGACAUAAUCCGUACACGGCCUUCCCUCCCGCAGUGCCCGGGCUGCCUCCGGGCCUCCCGCCGGCCGUCUCCUUUGGCUCCCUGCAGGGGGCCUUCCAGCCCAAGAGCACGAACCCUGAGCUGCCACCACGACUGGGGCCGGUGCCGAGCGGGCUCUCCCAGAAGGGGACACAGAUCCCCGACCAUUUCCGGCCACCUUUGAGGCUCUUCACGGCCGCCCCCCAAGGCCCCGGCCCCUCCCGUGGCUCAGCCUC | circ |
| ENSG00000156860:+:16:30664214:30666541 | ENSG00000156860 | ENST00000356166 | + | 16 | 30664215 | 30666541 | 22 | CACCUUUGAGGCUCUUCACGGC | bsj |
| ENSG00000156860:+:16:30664214:30666541 | ENSG00000156860 | ENST00000356166 | + | 16 | 30664015 | 30664224 | 210 | GGGAGAAAUAGGGGUUGCUGGGUGUGGGAGGCGCCUCAGUCUGAGAACGGAUUCCCUCGGUGGGGGGCGGUCCUGGGGCUGGGUUCUGCCACCUCCCGUGUUGGAGAACCAGGGUCACGGUAUCACCCAGGCUUCUGACCCCCUCCCGUCAGAGCUGGCACCUCCCAACCCCUCACUCAUCUCCCCACUUCUCUCCCCAGCUCUUCACGG | ie_up |
| ENSG00000156860:+:16:30664214:30666541 | ENSG00000156860 | ENST00000356166 | + | 16 | 30666532 | 30666741 | 210 | ACCUUUGAGGGUGAGUUGUGUGAGGACCUCAGGCUGCAUGAGGCUGGGGGCUGGUGUUAGCAUGUUUGGCUAAGGGGGGUUUUGCUUACAAAUAAGUGAGAAGCCCAGAACAUGGAGGCAGCCAGAUGUGGAACAGCAGAGAGUGAGGGCUUUCAAGUUGCAGACCUGGGCUCGGAUGGAACUGGCCAUGGGACCUUGGGCAAGGCAGUU | ie_down |
- Note:
- id: unique identifier.
- gene: represents the gene from which the circRNA arises.
- transcript: transcript whose exon coordinates overlap with the detected back-spliced junction coordinate and used in the downstream analysis.
- strand: is the strand from which the gene is transcribed.
- chrom: is the chromosome from which the circRNA is derived.
- startGR: is the 5’ coordinate of the genomic range from which the sequence are extracted.
- endGR: is the 3’ coordinate of the genomic range from which the sequence are extracted.
- length: length of the extracted sequences.
- seq: sequence used in the downstream analysis.
- type: type of sequences retrieved. If type = “circ” the sequences derive from the internal circRNA sequences. If type = “bsj” the sequences derive from the back spliced junctions (BSJ). If type = “ie_up” the Intron or Exon sequences derive from the up-stream of BSJ up to 210 bp. If type = “ie_down” the Intron or Exon sequences derive from the down-stream of BSJ up to 210 bp.
2. RNA Binding Proteins Analysis
RBP on full sequence
Plot
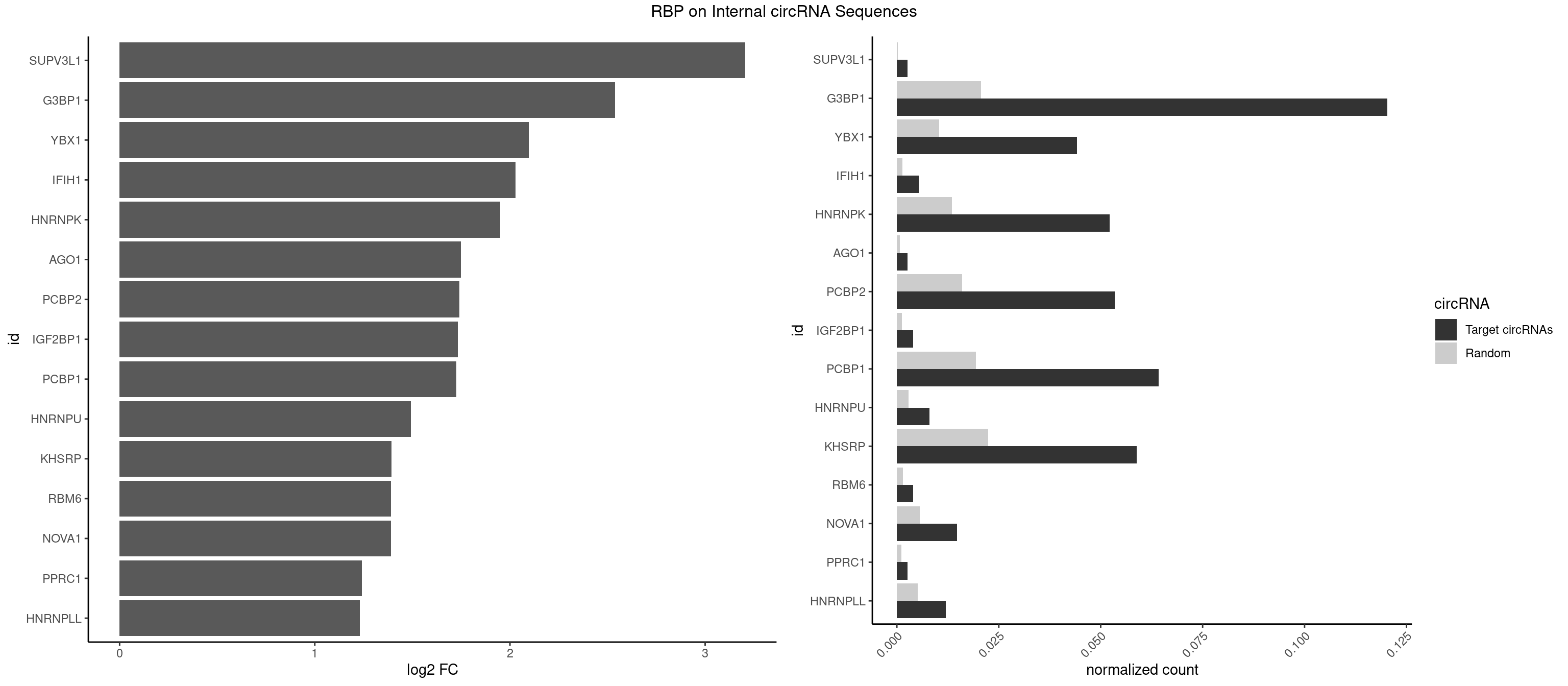
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| SUPV3L1 | 1 | 199 | 0.002673797 | 0.0002898408 | 3.205557 | CCGCCC | CCGCCC |
| G3BP1 | 89 | 14282 | 0.120320856 | 0.0206989801 | 2.539255 | ACCCAC,ACCCCC,ACCCCU,ACCGGC,ACGCCC,AGGCCC,AUCGGC,CACCGG,CCACAG,CCACCC,CCACCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCCAC,CCCCCA,CCCCCC,CCCCCG,CCCCGG,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUCC,CCGCAG,CCGCCC,CCGCCG,CCGGCC,CCUCCG,CGGCCC,CGGCCG,CUCGGC,UCGGCC | ACACGC,ACAGGC,ACCCAC,ACCCAU,ACCCCC,ACCCCU,ACCCGC,ACCGGC,ACGCAC,ACGCAG,ACGCCC,ACGCCG,AGGCAC,AGGCAG,AGGCCC,AGGCCG,AUACGC,AUAGGC,AUCCGC,AUCGGC,CACACG,CACAGG,CACCCG,CACCGG,CACGCA,CACGCC,CAGGCA,CAGGCC,CAUACG,CAUAGG,CAUCCG,CAUCGG,CCACAC,CCACAG,CCACCC,CCACCG,CCACGC,CCAGGC,CCAUAC,CCAUAG,CCAUCC,CCAUCG,CCCACA,CCCACC,CCCACG,CCCAGG,CCCAUA,CCCAUC,CCCCAC,CCCCAG,CCCCCA,CCCCCC,CCCCCG,CCCCGC,CCCCGG,CCCCUA,CCCCUC,CCCGCA,CCCGCC,CCCGGC,CCCUAC,CCCUAG,CCCUCC,CCCUCG,CCGCAC,CCGCAG,CCGCCC,CCGCCG,CCGGCA,CCGGCC,CCUACG,CCUAGG,CCUCCG,CCUCGG,CGGCAC,CGGCAG,CGGCCC,CGGCCG,CUACGC,CUAGGC,CUCCGC,CUCGGC,UACGCA,UACGCC,UAGGCA,UAGGCC,UCCGCA,UCCGCC,UCGGCA,UCGGCC |
| YBX1 | 32 | 7119 | 0.044117647 | 0.0103183321 | 2.096146 | AACCAC,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAUCAU,CAUCUG,CCACCA,CCAGCA,CCCUGC,CUGCGG,GCCUGC,UCCAGC | AACAUC,AACCAC,ACACCA,ACAUCA,ACAUCG,ACAUCU,ACCACA,ACCACC,AUCAUC,CAACCA,CACACC,CACCAC,CAGCAA,CAUCAU,CAUCGC,CAUCUG,CCACAA,CCACAC,CCACCA,CCAGCA,CCCUGC,CCUGCG,CUGCGG,GAUCUG,GCCUGC,GGUCUG,GUCUGC,UCCAGC,UGCGGU |
| IFIH1 | 3 | 904 | 0.005347594 | 0.0013115296 | 2.027639 | GGCCCU,GGCCGC,GGGCCG | CCGCGG,CGCGGA,GCCGCG,GCGGAU,GGCCCU,GGCCGC,GGGCCG |
| HNRNPK | 38 | 9304 | 0.052139037 | 0.0134848428 | 1.951025 | ACCCCA,ACCCCC,CCCCAA,CCCCAC,CCCCCA,CCCCCC,CCCCCU,CCCCUU,GCCCAA,GCCCAG,GCCCCC,GCCCCU,UCCCCC,UCCCCU | AAAAAA,ACCCAA,ACCCAU,ACCCCA,ACCCCC,ACCCGA,ACCCGC,ACCCGU,ACCCUU,CAAACC,CAACCC,CAUACC,CAUCCC,CCAAAC,CCAACC,CCAUAC,CCAUCC,CCCCAA,CCCCAC,CCCCAU,CCCCCA,CCCCCC,CCCCCU,CCCCUA,CCCCUU,GCCCAA,GCCCAC,GCCCAG,GCCCCC,GCCCCU,GGGGGG,UCCCAA,UCCCAC,UCCCAU,UCCCCA,UCCCCC,UCCCCU,UCCCGA,UCCCUA,UCCCUU,UUUUUU |
| AGO1 | 1 | 548 | 0.002673797 | 0.0007956130 | 1.748751 | UGAGGU | AGGUAG,GAGGUA,GGUAGU,GUAGUA,UGAGGU |
| PCBP2 | 39 | 11045 | 0.053475936 | 0.0160079069 | 1.740105 | AACCCU,CCCCCA,CCCCCC,CCCCCU,CCCUCC,CCCUUA,CCUCCC,CCUCCU,CCUUCC,CUCCCA,CUCCCC,CUCCCU,CUUCCA,CUUCCC | AAACCA,AAAUUA,AAAUUC,AACCAA,AACCCU,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUA,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAUUA,CAUUAA,CCAUUA,CCAUUC,CCCCCA,CCCCCC,CCCCCU,CCCUAA,CCCUCC,CCCUUA,CCCUUC,CCUCCA,CCUCCC,CCUCCU,CCUUAA,CCUUCC,CUAACC,CUCCCA,CUCCCC,CUCCCU,CUUAAA,CUUCCA,CUUCCC,CUUCCU,GGGGGG,UAACCC,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| IGF2BP1 | 2 | 831 | 0.004010695 | 0.0012057377 | 1.733936 | AGCACC,GCACCC | AAGCAC,ACCCGU,AGCACC,CACCCG,CCCGUU,GCACCC |
| PCBP1 | 47 | 13384 | 0.064171123 | 0.0193975949 | 1.726046 | CACCCU,CCACCC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUUA,CCUCCC,CCUCUU,CCUUCC,CCUUUC,CUUCCC,CUUUCC | AAAAAA,AAACCA,AAAUUA,AAAUUC,AACCAA,AAUUAA,AAUUAC,AAUUCA,AAUUCC,ACAUUA,ACCAAA,ACCCUC,ACCUUA,AUUAAA,AUUACC,AUUCAC,AUUCCA,AUUCCC,CAAACC,CAACCC,CAAUCC,CAAUUA,CACCCU,CAUACC,CAUCCC,CAUUAA,CAUUCC,CCAAAC,CCAACC,CCAAUC,CCACCC,CCAUAC,CCAUCC,CCAUUA,CCAUUC,CCCACC,CCCCAA,CCCCAC,CCCCCC,CCCUCC,CCCUCU,CCCUUA,CCCUUC,CCUAAC,CCUACC,CCUAUC,CCUCCC,CCUCUU,CCUUAA,CCUUAC,CCUUCC,CCUUUC,CUAACC,CUACCC,CUAUCC,CUUAAA,CUUACC,CUUCCC,CUUUCC,GGGGGG,UCCCCA,UUAGAG,UUAGGA,UUAGGG,UUUUUU |
| HNRNPU | 5 | 1965 | 0.008021390 | 0.0028491350 | 1.493328 | CCCCCC | AAAAAA,CCCCCC,GGGGGG,UUUUUU |
| KHSRP | 43 | 15466 | 0.058823529 | 0.0224148375 | 1.391939 | AGCCUC,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCUUCC,CGCCCU,CGGCCU,CUCCCU,CUCCUU,CUGCCU,GCCCUC,GCCUCC,GCCUUC,GGCCUC,UCCCUC,UGCCUC | ACCCUC,ACCUUC,AGCCUC,AGCUUC,AUAUUU,AUGUAU,AUGUGU,AUUAUU,AUUUAU,AUUUUA,CACCCU,CACCUU,CAGCCU,CAGCUU,CCCCCU,CCCCUC,CCCCUU,CCCUCC,CCCUUC,CCGCCU,CCGCUU,CCUUCC,CGCCCU,CGCCUC,CGCCUU,CGCUUC,CGGCCU,CGGCUU,CUCCCU,CUCCUU,CUGCCU,CUGCUU,CUGUAU,CUGUGU,GCCCUC,GCCUCC,GCCUUC,GCUUCC,GGCCUC,GGCUUC,GUAUAU,GUAUGU,GUGUAU,GUGUGU,UAGGUA,UAGGUU,UAGUAU,UAUAUU,UAUUAU,UAUUUA,UAUUUU,UCCCUC,UCCUUC,UGCAUG,UGCCUC,UGCUUC,UGUAUA,UGUAUG,UGUGUA,UGUGUG,UUAUUA,UUAUUU,UUGUAU,UUGUGU,UUUAUA,UUUAUU |
| RBM6 | 2 | 1054 | 0.004010695 | 0.0015289102 | 1.391349 | AUCCAG,UAUCCA | AAUCCA,AUCCAA,AUCCAG,CAUCCA,UAUCCA |
| NOVA1 | 10 | 3872 | 0.014705882 | 0.0056127669 | 1.389609 | ACCACC,AGCACC,AUCAUC,CCCCCC | AACACC,ACCACC,AGCACC,AGUCAC,AUCAAC,AUCACC,AUCAUC,AUUCAU,CAGUCA,CAUUCA,CCCCCC,GGGGGG,UCAGUC,UCAUUC,UUCAUU,UUUUUU |
| PPRC1 | 1 | 780 | 0.002673797 | 0.0011318283 | 1.240235 | CCGCGC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| HNRNPLL | 8 | 3534 | 0.012032086 | 0.0051229360 | 1.231844 | CACACC,CACCAC,CACUGC,GCACAC | ACAAAC,ACACAC,ACACCA,ACAUAC,ACCACA,ACCGCA,ACGACA,ACUGCA,AGACGA,CAAACA,CACACA,CACACC,CACCAC,CACCGC,CACUGC,CAGACG,CAUACA,GACGAC,GCAAAC,GCACAC,GCAUAC |
RBP on BSJ (Exon Only)
Plot
No results under this category.
Spreadsheet
No results under this category.
RBP on BSJ (Exon and Intron)
Plot
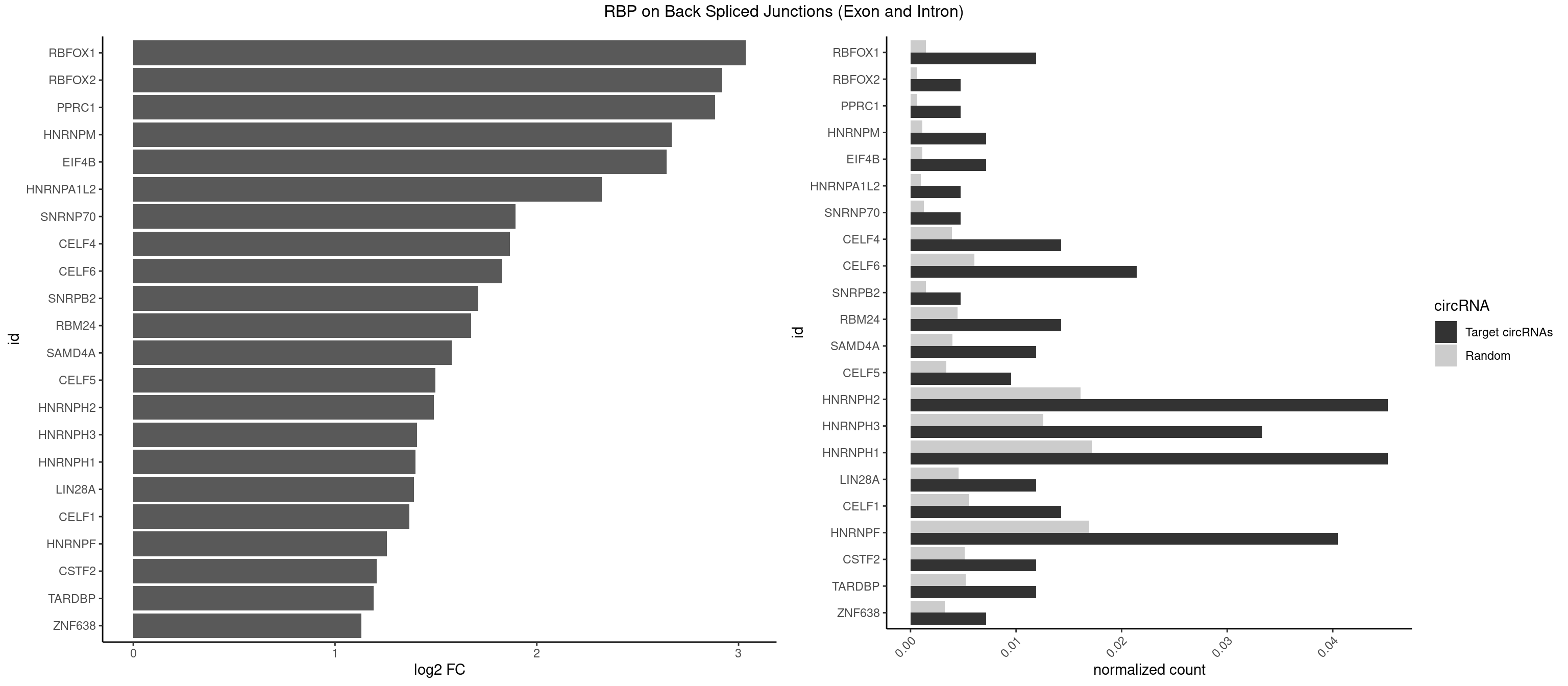
Spreadsheet
| id | foreground | background | foregroundNorm | backgroundNorm | log2FC | motifF | motifB |
|---|---|---|---|---|---|---|---|
| RBFOX1 | 4 | 287 | 0.011904762 | 0.0014510278 | 3.036392 | AGCAUG,GCAUGA,GCAUGU,UGCAUG | AGCAUG,GCAUGA,GCAUGC,GCAUGU,UGACUG,UGCAUG |
| RBFOX2 | 1 | 124 | 0.004761905 | 0.0006297864 | 2.918604 | UGCAUG | UGACUG,UGCAUG |
| PPRC1 | 1 | 127 | 0.004761905 | 0.0006449012 | 2.884389 | GGCGCC | CCGCGC,CGCGCC,CGCGCG,CGGCGC,GCGCGC,GGCGCC,GGCGCG,GGGCGC |
| HNRNPM | 2 | 222 | 0.007142857 | 0.0011235389 | 2.668451 | GGGGGG | AAGGAA,GAAGGA,GGGGGG |
| EIF4B | 2 | 226 | 0.007142857 | 0.0011436921 | 2.642803 | CUCGGA,GUUGGA | CUCGGA,CUUGGA,GUCGGA,GUUGGA,UCGGAA,UCGGAC,UUGGAA,UUGGAC |
| HNRNPA1L2 | 1 | 188 | 0.004761905 | 0.0009522370 | 2.322146 | AUAGGG | AUAGGG,GUAGGG,UAGGGA,UAGGGU,UUAGGG |
| SNRNP70 | 1 | 253 | 0.004761905 | 0.0012797259 | 1.895704 | UUCAAG | AAUCAA,AUCAAG,AUUCAA,GAUCAA,GUUCAA,UUCAAG |
| CELF4 | 5 | 776 | 0.014285714 | 0.0039147521 | 1.867580 | GGUGUG,GGUGUU,GUGUGG,GUGUUG,UGUGUG | GGUGUG,GGUGUU,GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| CELF6 | 8 | 1196 | 0.021428571 | 0.0060308343 | 1.829106 | GUGAGG,GUGGGG,GUGUGG,GUGUUG,UGUGAG,UGUGGG,UGUGUG | GUGAGG,GUGAUG,GUGGGG,GUGGUG,GUGUGG,GUGUUG,UGUGAG,UGUGAU,UGUGGG,UGUGGU,UGUGUG,UGUGUU |
| SNRPB2 | 1 | 288 | 0.004761905 | 0.0014560661 | 1.709463 | UUGCAG | AUUGCA,GUAUUG,UAUUGC,UGCAGU,UUGCAG |
| RBM24 | 5 | 888 | 0.014285714 | 0.0044790407 | 1.673311 | AGAGUG,GAGUGA,GUGUGA,GUGUGG,UGUGUG | AGAGUG,AGUGUG,GAGUGA,GAGUGG,GAGUGU,GUGUGA,GUGUGG,GUGUGU,UGAGUG,UGUGUG |
| SAMD4A | 4 | 790 | 0.011904762 | 0.0039852882 | 1.578783 | CUGGCA,CUGGCC,GCUGGC,GCUGGU | CGGGAA,CGGGAC,CGGGCA,CGGGCC,CGGGUA,CGGGUC,CUGGAA,CUGGAC,CUGGCA,CUGGCC,CUGGUA,CUGGUC,GCGGGA,GCGGGC,GCGGGU,GCUGGA,GCUGGC,GCUGGU |
| CELF5 | 3 | 669 | 0.009523810 | 0.0033756550 | 1.496371 | GUGUGG,GUGUUG,UGUGUG | GUGUGG,GUGUGU,GUGUUG,GUGUUU,UGUGUG,UGUGUU |
| HNRNPH2 | 18 | 3198 | 0.045238095 | 0.0161174929 | 1.488911 | AAGGGG,AUGUGG,CUGGGG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,GGGGGG,GGGUGU,UGGGGG,UGGGUG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGA,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH3 | 13 | 2496 | 0.033333333 | 0.0125806127 | 1.405763 | AUGUGG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,GGGGGG,GGGUGU,UGGGUG,UGUGGG | AAGGGA,AAGGUG,AAUGUG,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CGAGGG,CGGGCG,CGGGGG,GAAGGG,GAAUGU,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGUG,UGUGGG,UUGGGU |
| HNRNPH1 | 18 | 3406 | 0.045238095 | 0.0171654575 | 1.398030 | AAGGGG,AUGUGG,CUGGGG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,GGGGGG,GGGUGU,UGGGGG,UGGGUG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGGA,AUCGGG,AUGUGG,AUUGGG,CAGGAC,CGAGGG,CGGGCG,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGCGU,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGUCG,GGGUGU,GGUCGA,GUCGAG,UCGAGG,UCGGGC,UGGGGG,UGGGGU,UGGGUG,UGUGGG,UUGGGU |
| LIN28A | 4 | 900 | 0.011904762 | 0.0045395002 | 1.390933 | GGAGAA,UGGAGA,UGGAGG | AGGAGA,AGGAGU,CGGAGA,CGGAGG,CGGAGU,GGAGAA,GGAGAU,GGAGGA,GGAGGG,GGAGUA,UGGAGA,UGGAGG,UGGAGU |
| CELF1 | 5 | 1097 | 0.014285714 | 0.0055320435 | 1.368689 | GUGUUG,GUUGUG,UGUGUG,UGUUUG,UUGUGU | CUGUCU,GUGUGU,GUGUUG,GUUGUG,GUUUGU,UGUCUG,UGUGUG,UGUGUU,UGUUGU,UGUUUG,UUGUGU |
| HNRNPF | 16 | 3360 | 0.040476190 | 0.0169336961 | 1.257177 | AAGGGG,AUGGGA,AUGUGG,CUGGGG,GGAUGG,GGGAGG,GGGCUG,GGGGCU,GGGGGC,GGGGGG,UGUGGG | AAGGCG,AAGGGA,AAGGGG,AAGGUG,AAUGUG,AGGAAG,AGGCGA,AGGGAA,AGGGAU,AGGGGA,AUGGGA,AUGGGG,AUGUGG,CGAUGG,CGGGAU,CGGGGG,CUGGGG,GAAGGG,GAAGGU,GAAUGU,GAGGAA,GAGGGG,GAUGGG,GGAAGG,GGAAUG,GGAGGA,GGAGGG,GGAUGG,GGCGAA,GGGAAG,GGGAAU,GGGAGG,GGGAUG,GGGCUG,GGGGAA,GGGGAG,GGGGCU,GGGGGC,GGGGGG,GGGGUA,UGGGAA,UGGGGU,UGUGGG |
| CSTF2 | 4 | 1022 | 0.011904762 | 0.0051541717 | 1.207726 | GUGUUG,GUUUUG,UGUGUG,UGUUUG | GUGUGU,GUGUUG,GUGUUU,GUUUUG,UGUGUG,UGUGUU,UGUUUG,UGUUUU |
| TARDBP | 4 | 1034 | 0.011904762 | 0.0052146312 | 1.190902 | GUUGUG,GUUUUG,UGUGUG,UUUUGC | GAAUGA,GAAUGG,GAAUGU,GUGAAU,GUGUGU,GUUGUG,GUUGUU,GUUUUG,UGAAUG,UGUGUG,UUGUGC,UUGUUC,UUUUGC |
| ZNF638 | 2 | 646 | 0.007142857 | 0.0032597743 | 1.131729 | GGUUCU,UGUUGG | CGUUCG,CGUUCU,CGUUGG,CGUUGU,GGUUCG,GGUUCU,GGUUGG,GGUUGU,GUUCGU,GUUCUU,GUUGGU,GUUGUU,UGUUCG,UGUUCU,UGUUGG,UGUUGU |
Help
- Note:
- foreground: number of motifs found in the foreground target sequences (e.g. predicted circRNAs).
- background: number of motifs found in the background target sequences (e.g. randomly generated circRNAs).
- foregroundNorm: number of motifs found in the foreground target sequences (+1) divided by the number (or length) of sequences analyzed.
- backgroundNorm: number of motifs found in background target sequences (+1) divided by the number (or length) of sequences analyzed.
- log2FC: log2 fold change calculated in this way (foregroundNorm)/(backgroundNorm).
- motifF: motifs of the corresponding RBP found in the foreground sequences.
- motifB: motifs of the corresponding RBP found in the background sequences.