Isoforom Switch Analysis
WANG Ziyi
Date: 19 5月 2022
ENSG00000076555
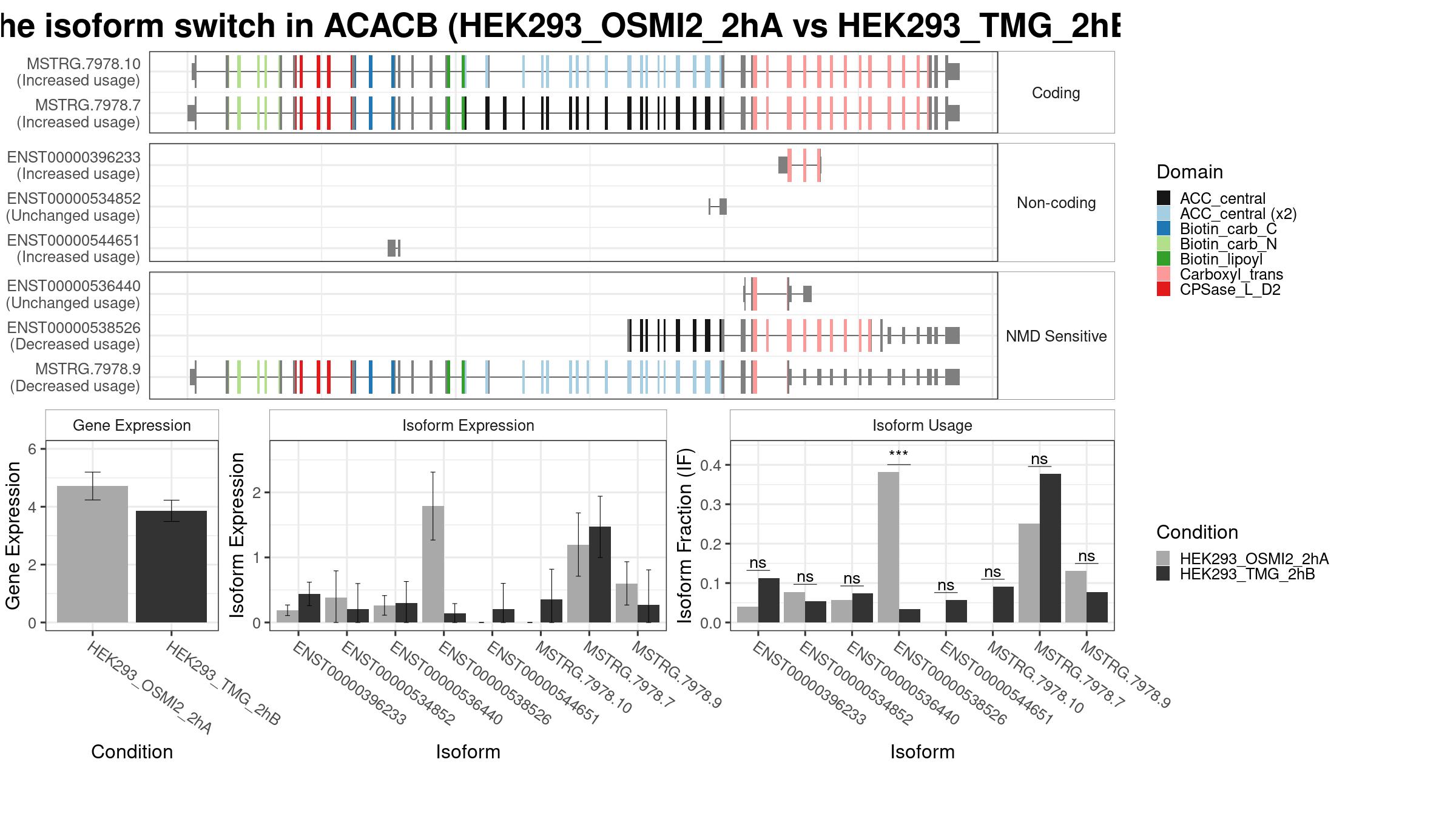
1 Isoform Switch
Summary Plot
Only the isoforms with fraction > 5% are shown below.
Notes on IDR and IDR_w_binding: As the prediction of Intrinsically Disordered Regions (IDR) and Intrinsically Disordered Binding Regions (IDBR) are done by two different tools we require two things to annotate the Intrinsically Disordered Binding Regions (IDBR). Firstly the IDBR (predicted by ANCHOR2) must be a region of at least 15 amino acids (after the smoothing). Secondly the fraction of the IDBR which overlaps the IDR predictions (done by IUPred3, again after smoothing) must be at least 80%. When that is the case the IDR type will be annotated as “IDR_w_binding_region” instead of just “IDR”. The current default parameters have not been rigorously tested and should be considered experimental.
For the function of any other domain, please Click HERE to search it through Pfam database.
Results
| isoform_id | gene_id | condition_1 | condition_2 | gene_name | gene_biotype | iso_biotype | gene_overall_mean | gene_value_1 | gene_value_2 | gene_stderr_1 | gene_stderr_2 | gene_log2_fold_change | iso_overall_mean | iso_value_1 | iso_value_2 | iso_stderr_1 | iso_stderr_2 | iso_log2_fold_change | IF_overall | IF1 | IF2 | dIF | isoform_switch_q_value | gene_switch_q_value | PTC | codingPotential |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENST00000396233 | ENSG00000076555 | HEK293_OSMI2_2hA | HEK293_TMG_2hB | ACACB | protein_coding | retained_intron | 4.15181 | 4.715539 | 3.858692 | 0.2452197 | 0.1877023 | -0.2886331 | 0.4005759 | 0.1876572 | 0.4387745 | 0.04169204 | 0.09200977 | 1.1829903 | 0.09711250 | 0.04046667 | 0.11306667 | 0.07260000 | 0.06152714 | 0.0004505 | FALSE | FALSE |
| ENST00000536440 | ENSG00000076555 | HEK293_OSMI2_2hA | HEK293_TMG_2hB | ACACB | protein_coding | nonsense_mediated_decay | 4.15181 | 4.715539 | 3.858692 | 0.2452197 | 0.1877023 | -0.2886331 | 0.2228168 | 0.2625367 | 0.2973469 | 0.07684920 | 0.16975351 | 0.1734176 | 0.05066250 | 0.05760000 | 0.07353333 | 0.01593333 | 0.95725304 | 0.0004505 | TRUE | TRUE |
| ENST00000538526 | ENSG00000076555 | HEK293_OSMI2_2hA | HEK293_TMG_2hB | ACACB | protein_coding | nonsense_mediated_decay | 4.15181 | 4.715539 | 3.858692 | 0.2452197 | 0.1877023 | -0.2886331 | 0.9672861 | 1.7898732 | 0.1400449 | 0.26628013 | 0.07649213 | -3.5844287 | 0.21177917 | 0.38150000 | 0.03463333 | -0.34686667 | 0.00045050 | 0.0004505 | TRUE | TRUE |
| ENST00000544651 | ENSG00000076555 | HEK293_OSMI2_2hA | HEK293_TMG_2hB | ACACB | protein_coding | retained_intron | 4.15181 | 4.715539 | 3.858692 | 0.2452197 | 0.1877023 | -0.2886331 | 0.1088520 | 0.0000000 | 0.2030242 | 0.00000000 | 0.20302422 | 4.4129456 | 0.02200000 | 0.00000000 | 0.05676667 | 0.05676667 | 0.79949725 | 0.0004505 | FALSE | |
| MSTRG.7978.10 | ENSG00000076555 | HEK293_OSMI2_2hA | HEK293_TMG_2hB | ACACB | protein_coding | 4.15181 | 4.715539 | 3.858692 | 0.2452197 | 0.1877023 | -0.2886331 | 0.2519843 | 0.0000000 | 0.3515379 | 0.00000000 | 0.23842480 | 5.1760749 | 0.07487083 | 0.00000000 | 0.09063333 | 0.09063333 | 0.19552013 | 0.0004505 | FALSE | TRUE | |
| MSTRG.7978.7 | ENSG00000076555 | HEK293_OSMI2_2hA | HEK293_TMG_2hB | ACACB | protein_coding | 4.15181 | 4.715539 | 3.858692 | 0.2452197 | 0.1877023 | -0.2886331 | 1.2476882 | 1.1978102 | 1.4697728 | 0.24829935 | 0.24038996 | 0.2929819 | 0.30777917 | 0.25100000 | 0.37676667 | 0.12576667 | 0.33141723 | 0.0004505 | FALSE | TRUE | |
| MSTRG.7978.9 | ENSG00000076555 | HEK293_OSMI2_2hA | HEK293_TMG_2hB | ACACB | protein_coding | 4.15181 | 4.715539 | 3.858692 | 0.2452197 | 0.1877023 | -0.2886331 | 0.5693540 | 0.6009359 | 0.2732347 | 0.16977801 | 0.27323471 | -1.1090230 | 0.14262500 | 0.13063333 | 0.07640000 | -0.05423333 | 0.35408499 | 0.0004505 | TRUE | TRUE |
- Note:
- The comparisons made can be identified as “from ‘condition_1’ to ‘condition_2’”, meaning ‘condition_1’ is considered the ground state and ‘condition_2’ the changed state. This also means that a positive dIF value indicates that the isoform usage is increased in ‘condition_2’ compared to ‘condition_1’. Since the ‘isoformFeatures’ entry is the most relevant part of the switchAnalyzeRlist object, the most-used standard methods have also been implemented to work directly on isoformFeatures.
2 Differential Exon Usage
Counts
All exons whithin this gene region are shown and numbering below.
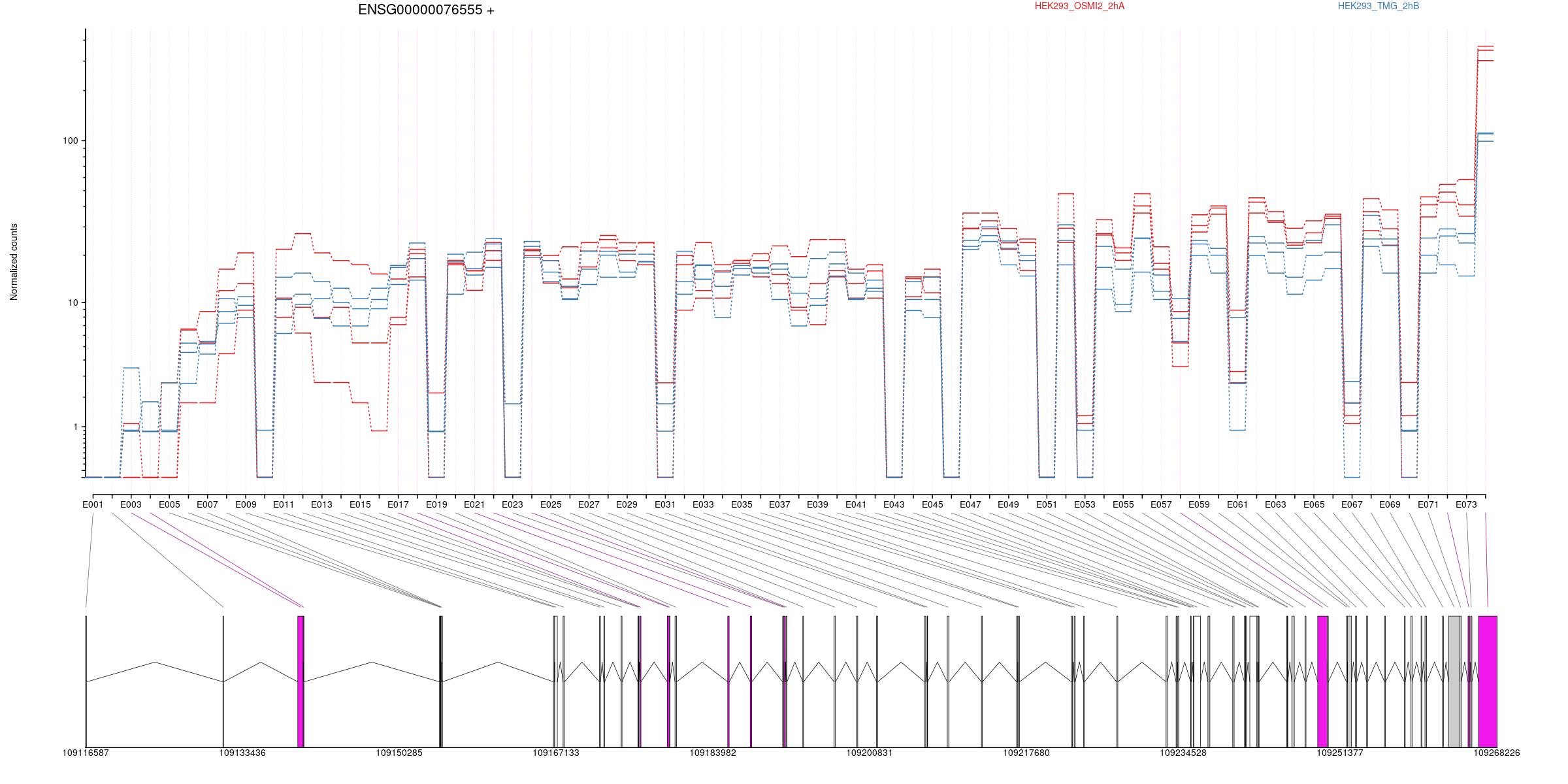
Expression
All exons whithin this gene region are shown and numbering below.
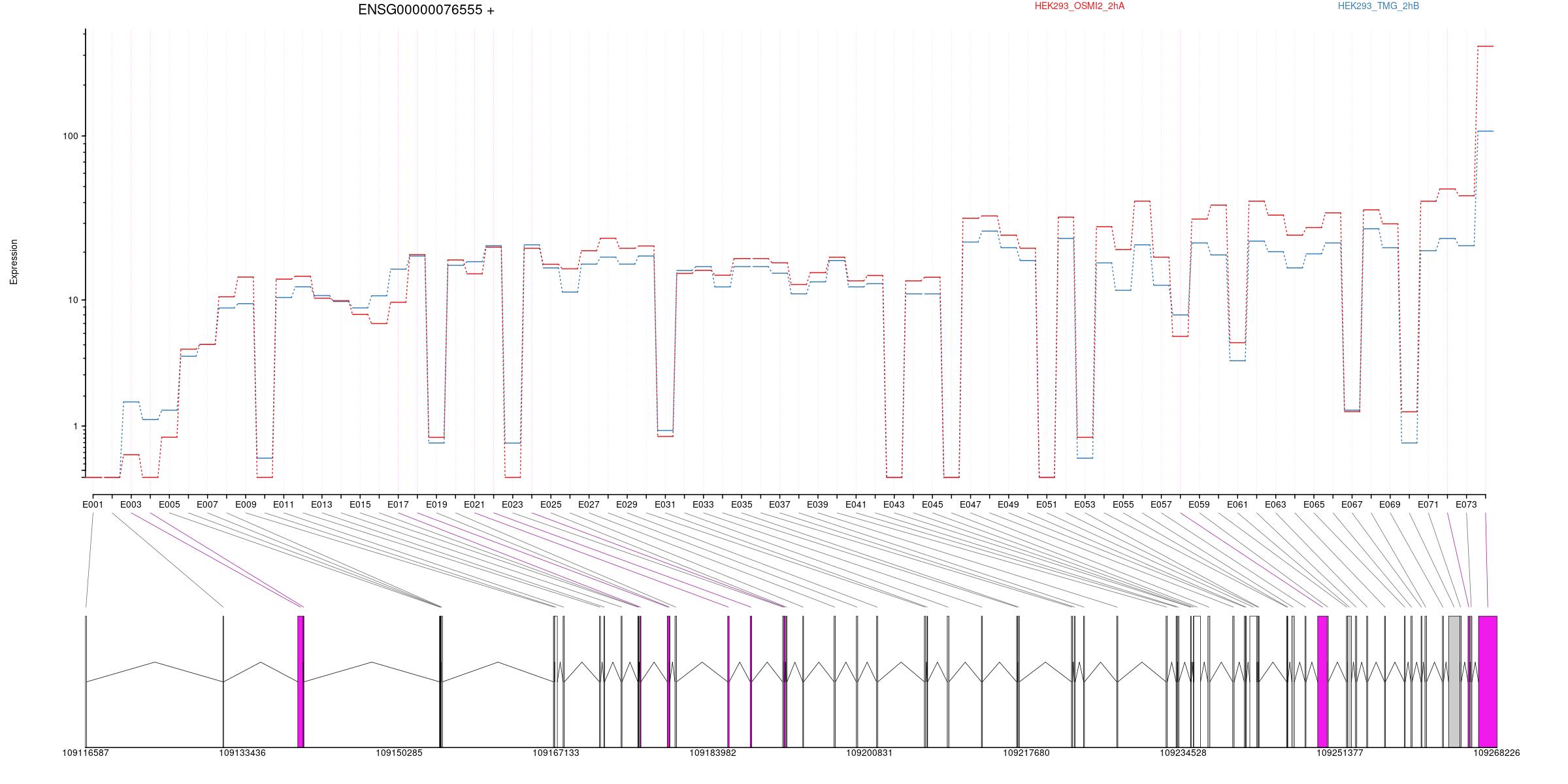
Splicing
All exons whithin this gene region are shown and numbering below.
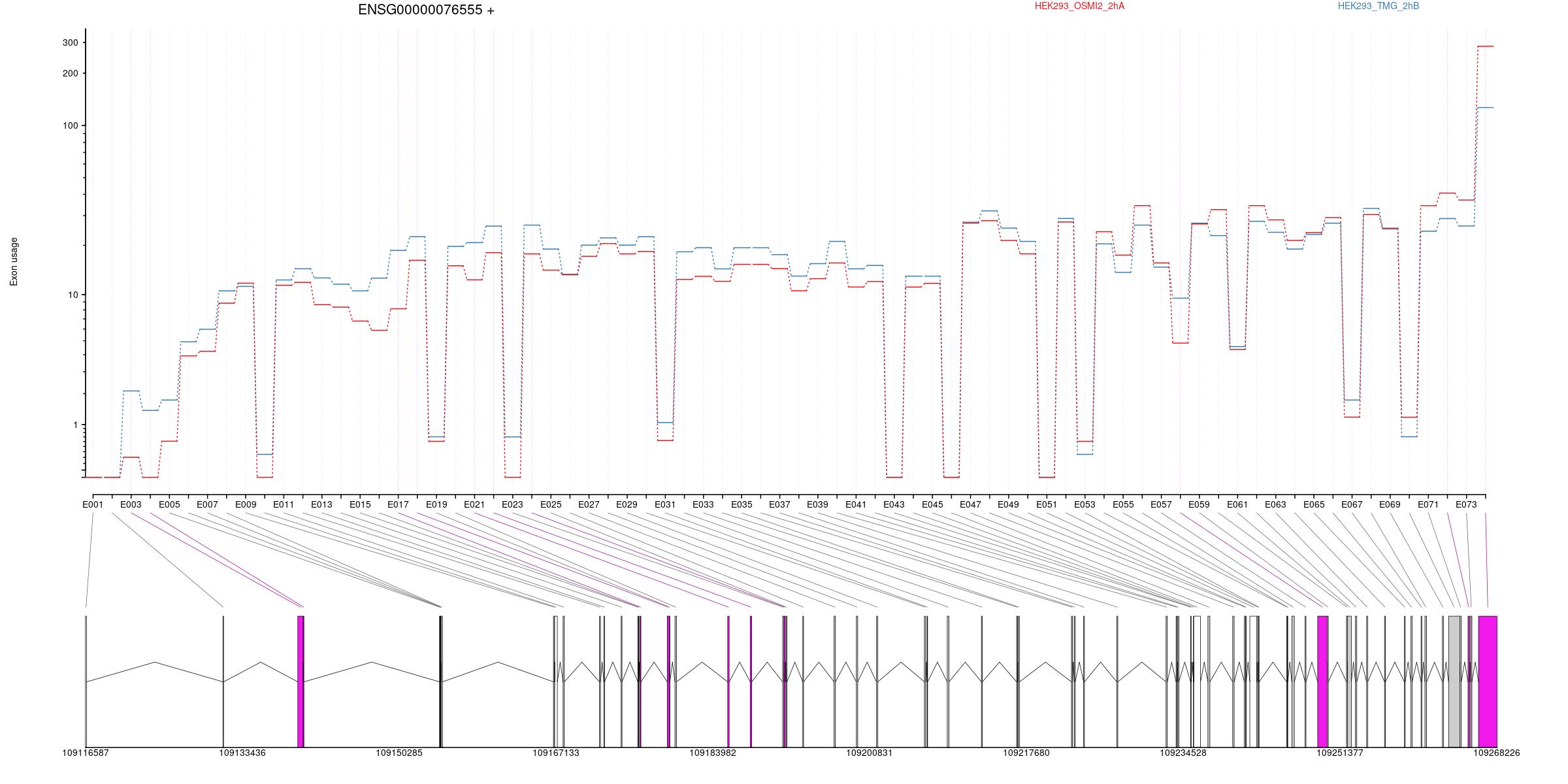
Transcripts
All isoforms whithin this gene region are shown below.
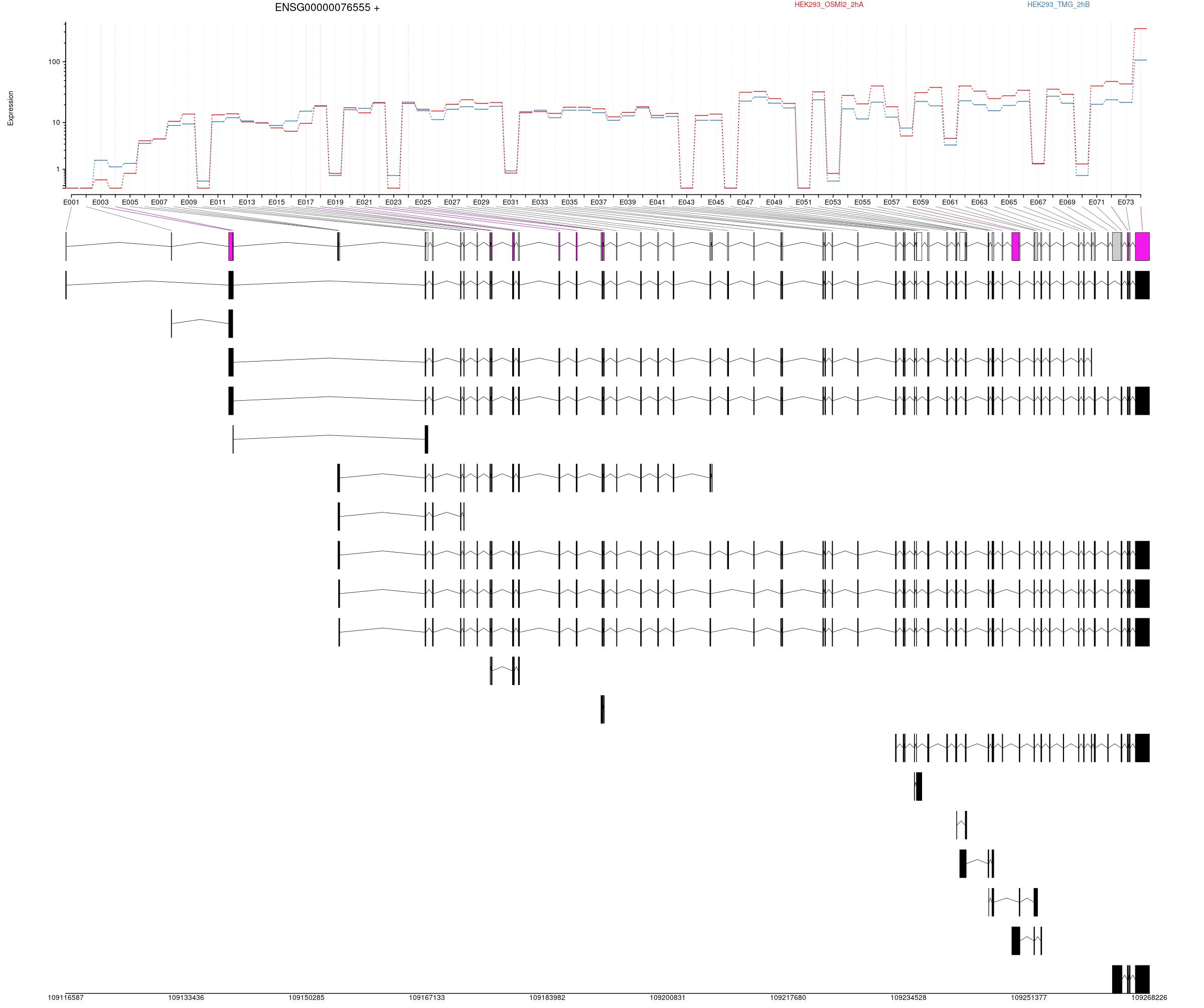
Results
| groupID | featureID | exonBaseMean | dispersion | pvalue | padj | seqnames | start | end | width | strand | HEK293_TMG_2hB | HEK293_OSMI2_2hA | log2fold_HEK293_OSMI2_2hA_HEK293_TMG_2hB |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ENSG00000076555 | E001 | 0.0000000 | 12 | 109116587 | 109116704 | 118 | + | ||||||
| ENSG00000076555 | E002 | 0.0000000 | 12 | 109131358 | 109131426 | 69 | + | ||||||
| ENSG00000076555 | E003 | 1.0611596 | 0.0894102418 | 3.942522e-02 | 8.996723e-02 | 12 | 109139397 | 109139978 | 582 | + | 0.492 | 0.114 | -2.805 |
| ENSG00000076555 | E004 | 0.5954526 | 0.0183557928 | 2.252660e-02 | 5.677486e-02 | 12 | 109139979 | 109140058 | 80 | + | 0.381 | 0.000 | -13.031 |
| ENSG00000076555 | E005 | 1.1813256 | 0.0495058410 | 1.814654e-01 | 3.030855e-01 | 12 | 109154623 | 109154681 | 59 | + | 0.441 | 0.206 | -1.535 |
| ENSG00000076555 | E006 | 4.5706280 | 0.0046175602 | 4.715642e-01 | 6.106759e-01 | 12 | 109154682 | 109154756 | 75 | + | 0.773 | 0.692 | -0.328 |
| ENSG00000076555 | E007 | 5.1523760 | 0.0089144770 | 2.807024e-01 | 4.205774e-01 | 12 | 109154757 | 109154793 | 37 | + | 0.844 | 0.718 | -0.500 |
| ENSG00000076555 | E008 | 9.8908771 | 0.0220515208 | 4.279371e-01 | 5.707646e-01 | 12 | 109154794 | 109154940 | 147 | + | 1.062 | 0.992 | -0.257 |
| ENSG00000076555 | E009 | 11.8900930 | 0.0063768876 | 9.564565e-01 | 9.767234e-01 | 12 | 109166861 | 109166993 | 133 | + | 1.088 | 1.106 | 0.065 |
| ENSG00000076555 | E010 | 0.1515154 | 0.0428844150 | 4.026043e-01 | 12 | 109166994 | 109167280 | 287 | + | 0.131 | 0.000 | -10.946 | |
| ENSG00000076555 | E011 | 11.9393393 | 0.0451256052 | 6.517509e-01 | 7.625604e-01 | 12 | 109167896 | 109168034 | 139 | + | 1.125 | 1.095 | -0.108 |
| ENSG00000076555 | E012 | 13.2135247 | 0.0779034273 | 3.799563e-01 | 5.242307e-01 | 12 | 109171805 | 109171914 | 110 | + | 1.189 | 1.111 | -0.278 |
| ENSG00000076555 | E013 | 10.5722079 | 0.1037391020 | 2.159448e-01 | 3.455390e-01 | 12 | 109172275 | 109172356 | 82 | + | 1.136 | 0.984 | -0.554 |
| ENSG00000076555 | E014 | 9.9735080 | 0.0778057345 | 2.653054e-01 | 4.033425e-01 | 12 | 109174132 | 109174230 | 99 | + | 1.101 | 0.970 | -0.479 |
| ENSG00000076555 | E015 | 8.5331613 | 0.1189732292 | 1.883253e-01 | 3.116230e-01 | 12 | 109175931 | 109175976 | 46 | + | 1.063 | 0.891 | -0.640 |
| ENSG00000076555 | E016 | 8.8966482 | 0.1345229901 | 5.612845e-02 | 1.199288e-01 | 12 | 109175977 | 109176040 | 64 | + | 1.135 | 0.838 | -1.104 |
| ENSG00000076555 | E017 | 12.7312885 | 0.0013611830 | 1.338500e-04 | 6.759834e-04 | 12 | 109176153 | 109176263 | 111 | + | 1.293 | 0.961 | -1.196 |
| ENSG00000076555 | E018 | 18.9610387 | 0.0011040325 | 4.174743e-02 | 9.427060e-02 | 12 | 109179088 | 109179297 | 210 | + | 1.371 | 1.237 | -0.469 |
| ENSG00000076555 | E019 | 0.6558305 | 0.0557978190 | 8.343455e-01 | 8.962794e-01 | 12 | 109179298 | 109179371 | 74 | + | 0.231 | 0.205 | -0.221 |
| ENSG00000076555 | E020 | 17.2776476 | 0.0010540163 | 1.002277e-01 | 1.908256e-01 | 12 | 109179917 | 109180087 | 171 | + | 1.316 | 1.206 | -0.387 |
| ENSG00000076555 | E021 | 16.0717372 | 0.0012703602 | 4.872986e-03 | 1.557650e-02 | 12 | 109185579 | 109185740 | 162 | + | 1.337 | 1.125 | -0.749 |
| ENSG00000076555 | E022 | 21.6165916 | 0.0022411302 | 1.901505e-02 | 4.936613e-02 | 12 | 109187999 | 109188162 | 164 | + | 1.432 | 1.280 | -0.526 |
| ENSG00000076555 | E023 | 0.2903454 | 0.4545930252 | 2.493211e-01 | 12 | 109191478 | 109191612 | 135 | + | 0.230 | 0.000 | -10.862 | |
| ENSG00000076555 | E024 | 21.6070016 | 0.0009482466 | 1.009238e-02 | 2.898947e-02 | 12 | 109191613 | 109191763 | 151 | + | 1.437 | 1.273 | -0.570 |
| ENSG00000076555 | E025 | 16.6131196 | 0.0011500928 | 8.383487e-02 | 1.655541e-01 | 12 | 109191847 | 109191950 | 104 | + | 1.301 | 1.181 | -0.423 |
| ENSG00000076555 | E026 | 13.8331065 | 0.0014608718 | 7.803495e-01 | 8.588368e-01 | 12 | 109193648 | 109193729 | 82 | + | 1.157 | 1.154 | -0.011 |
| ENSG00000076555 | E027 | 18.7970347 | 0.0010354618 | 2.687974e-01 | 4.072721e-01 | 12 | 109197008 | 109197153 | 146 | + | 1.323 | 1.259 | -0.223 |
| ENSG00000076555 | E028 | 21.6126297 | 0.0009049257 | 4.495172e-01 | 5.907180e-01 | 12 | 109199402 | 109199552 | 151 | + | 1.365 | 1.332 | -0.113 |
| ENSG00000076555 | E029 | 19.0581073 | 0.0009856912 | 3.504939e-01 | 4.946661e-01 | 12 | 109201567 | 109201701 | 135 | + | 1.323 | 1.274 | -0.173 |
| ENSG00000076555 | E030 | 20.3628623 | 0.0009947231 | 1.538408e-01 | 2.668927e-01 | 12 | 109206710 | 109206856 | 147 | + | 1.371 | 1.287 | -0.293 |
| ENSG00000076555 | E031 | 0.8804848 | 0.2345653091 | 5.655701e-01 | 6.926461e-01 | 12 | 109207002 | 109207057 | 56 | + | 0.312 | 0.210 | -0.761 |
| ENSG00000076555 | E032 | 15.4058329 | 0.0093527184 | 6.088505e-02 | 1.281240e-01 | 12 | 109209165 | 109209353 | 189 | + | 1.285 | 1.128 | -0.558 |
| ENSG00000076555 | E033 | 15.9082527 | 0.0122840732 | 5.095308e-02 | 1.108612e-01 | 12 | 109212836 | 109212936 | 101 | + | 1.308 | 1.146 | -0.575 |
| ENSG00000076555 | E034 | 13.4036964 | 0.0108734208 | 3.486321e-01 | 4.928609e-01 | 12 | 109216618 | 109216706 | 89 | + | 1.188 | 1.117 | -0.254 |
| ENSG00000076555 | E035 | 17.2953786 | 0.0010609858 | 1.457081e-01 | 2.558125e-01 | 12 | 109216796 | 109216920 | 125 | + | 1.308 | 1.214 | -0.332 |
| ENSG00000076555 | E036 | 17.0673897 | 0.0010849032 | 1.451539e-01 | 2.550475e-01 | 12 | 109222507 | 109222620 | 114 | + | 1.308 | 1.214 | -0.333 |
| ENSG00000076555 | E037 | 15.9655585 | 0.0012851252 | 2.204830e-01 | 3.509893e-01 | 12 | 109222799 | 109222912 | 114 | + | 1.269 | 1.189 | -0.282 |
| ENSG00000076555 | E038 | 11.8035927 | 0.0161524771 | 3.304511e-01 | 4.740350e-01 | 12 | 109223815 | 109223904 | 90 | + | 1.147 | 1.063 | -0.302 |
| ENSG00000076555 | E039 | 14.1257100 | 0.0541412174 | 3.839772e-01 | 5.281762e-01 | 12 | 109227371 | 109227489 | 119 | + | 1.218 | 1.132 | -0.305 |
| ENSG00000076555 | E040 | 18.1821463 | 0.0011096248 | 6.486086e-02 | 1.348083e-01 | 12 | 109232669 | 109232806 | 138 | + | 1.344 | 1.222 | -0.430 |
| ENSG00000076555 | E041 | 12.8043587 | 0.0013939398 | 1.727416e-01 | 2.917662e-01 | 12 | 109233748 | 109233847 | 100 | + | 1.188 | 1.084 | -0.371 |
| ENSG00000076555 | E042 | 13.6980807 | 0.0013051711 | 2.039233e-01 | 3.310992e-01 | 12 | 109233938 | 109234045 | 108 | + | 1.207 | 1.116 | -0.327 |
| ENSG00000076555 | E043 | 0.0000000 | 12 | 109235307 | 109235312 | 6 | + | ||||||
| ENSG00000076555 | E044 | 12.1129259 | 0.0015742945 | 3.725469e-01 | 5.169898e-01 | 12 | 109235313 | 109235369 | 57 | + | 1.146 | 1.084 | -0.223 |
| ENSG00000076555 | E045 | 12.5808346 | 0.0015794709 | 4.986803e-01 | 6.351158e-01 | 12 | 109235606 | 109235647 | 42 | + | 1.146 | 1.106 | -0.147 |
| ENSG00000076555 | E046 | 0.0000000 | 12 | 109235648 | 109236377 | 730 | + | ||||||
| ENSG00000076555 | E047 | 27.3980088 | 0.0008775025 | 6.653724e-01 | 7.730598e-01 | 12 | 109237165 | 109237380 | 216 | + | 1.454 | 1.449 | -0.017 |
| ENSG00000076555 | E048 | 29.8852615 | 0.0007253700 | 2.043620e-01 | 3.316155e-01 | 12 | 109239830 | 109239985 | 156 | + | 1.518 | 1.462 | -0.192 |
| ENSG00000076555 | E049 | 23.1893722 | 0.0011129306 | 1.865925e-01 | 3.094634e-01 | 12 | 109241078 | 109241222 | 145 | + | 1.420 | 1.350 | -0.244 |
| ENSG00000076555 | E050 | 19.7677371 | 0.0009901566 | 2.223655e-01 | 3.532203e-01 | 12 | 109241223 | 109241281 | 59 | + | 1.344 | 1.274 | -0.247 |
| ENSG00000076555 | E051 | 0.0000000 | 12 | 109241670 | 109242436 | 767 | + | ||||||
| ENSG00000076555 | E052 | 29.0599125 | 0.0161045077 | 6.173893e-01 | 7.352127e-01 | 12 | 109242437 | 109242592 | 156 | + | 1.475 | 1.455 | -0.069 |
| ENSG00000076555 | E053 | 0.5546650 | 0.0205046804 | 6.935743e-01 | 7.946031e-01 | 12 | 109242593 | 109242667 | 75 | + | 0.131 | 0.205 | 0.779 |
| ENSG00000076555 | E054 | 23.1455698 | 0.0009432462 | 5.204118e-01 | 6.543771e-01 | 12 | 109245626 | 109245713 | 88 | + | 1.330 | 1.400 | 0.241 |
| ENSG00000076555 | E055 | 16.0586895 | 0.0015474510 | 3.749176e-01 | 5.193552e-01 | 12 | 109245714 | 109245748 | 35 | + | 1.168 | 1.266 | 0.349 |
| ENSG00000076555 | E056 | 31.9008720 | 0.0006686515 | 1.601128e-01 | 2.752705e-01 | 12 | 109246179 | 109246448 | 270 | + | 1.437 | 1.548 | 0.381 |
| ENSG00000076555 | E057 | 15.6710041 | 0.0012109859 | 9.851816e-01 | 9.947486e-01 | 12 | 109247606 | 109247703 | 98 | + | 1.198 | 1.222 | 0.085 |
| ENSG00000076555 | E058 | 6.9152237 | 0.0034499735 | 2.021710e-02 | 5.194214e-02 | 12 | 109248958 | 109249983 | 1026 | + | 1.021 | 0.765 | -0.977 |
| ENSG00000076555 | E059 | 27.0457739 | 0.0010570841 | 6.751259e-01 | 7.804867e-01 | 12 | 109249984 | 109250104 | 121 | + | 1.448 | 1.444 | -0.015 |
| ENSG00000076555 | E060 | 28.8229424 | 0.0007277188 | 5.802614e-02 | 1.232524e-01 | 12 | 109252046 | 109252156 | 111 | + | 1.378 | 1.525 | 0.509 |
| ENSG00000076555 | E061 | 4.3832101 | 0.1658596429 | 9.224702e-01 | 9.550937e-01 | 12 | 109252157 | 109252578 | 422 | + | 0.745 | 0.729 | -0.066 |
| ENSG00000076555 | E062 | 32.3598720 | 0.0006617011 | 2.867112e-01 | 4.271301e-01 | 12 | 109253015 | 109253158 | 144 | + | 1.459 | 1.548 | 0.306 |
| ENSG00000076555 | E063 | 27.0257822 | 0.0007899735 | 4.783265e-01 | 6.167520e-01 | 12 | 109254214 | 109254334 | 121 | + | 1.396 | 1.467 | 0.244 |
| ENSG00000076555 | E064 | 20.7403721 | 0.0010183582 | 7.269882e-01 | 8.198693e-01 | 12 | 109256140 | 109256236 | 97 | + | 1.301 | 1.350 | 0.172 |
| ENSG00000076555 | E065 | 23.8133088 | 0.0008769991 | 8.562465e-01 | 9.111163e-01 | 12 | 109258268 | 109258364 | 97 | + | 1.384 | 1.394 | 0.036 |
| ENSG00000076555 | E066 | 28.8255534 | 0.0008532206 | 8.910874e-01 | 9.344542e-01 | 12 | 109258973 | 109259108 | 136 | + | 1.448 | 1.480 | 0.110 |
| ENSG00000076555 | E067 | 1.4487803 | 0.0106287276 | 5.237893e-01 | 6.574200e-01 | 12 | 109260065 | 109260149 | 85 | + | 0.441 | 0.343 | -0.546 |
| ENSG00000076555 | E068 | 32.2835818 | 0.0018828004 | 3.588341e-01 | 5.032167e-01 | 12 | 109260480 | 109260657 | 178 | + | 1.532 | 1.498 | -0.118 |
| ENSG00000076555 | E069 | 25.7013155 | 0.0088106871 | 7.174226e-01 | 8.128024e-01 | 12 | 109262357 | 109262469 | 113 | + | 1.420 | 1.416 | -0.012 |
| ENSG00000076555 | E070 | 0.9650892 | 0.0361997596 | 5.843549e-01 | 7.083170e-01 | 12 | 109263022 | 109264231 | 1210 | + | 0.231 | 0.342 | 0.771 |
| ENSG00000076555 | E071 | 30.3956015 | 0.0010919736 | 5.870793e-02 | 1.244174e-01 | 12 | 109264232 | 109264386 | 155 | + | 1.402 | 1.548 | 0.501 |
| ENSG00000076555 | E072 | 36.4784682 | 0.0010367652 | 4.111997e-02 | 9.310983e-02 | 12 | 109265110 | 109265280 | 171 | + | 1.475 | 1.620 | 0.496 |
| ENSG00000076555 | E073 | 33.3687377 | 0.0123004668 | 1.188624e-01 | 2.183169e-01 | 12 | 109265389 | 109265525 | 137 | + | 1.432 | 1.580 | 0.508 |
| ENSG00000076555 | E074 | 223.0914998 | 0.0065360053 | 1.065428e-14 | 3.488458e-13 | 12 | 109266236 | 109268226 | 1991 | + | 2.107 | 2.457 | 1.167 |